5YRX
 
 | | Crystal structure of a hypothetical protein Rv3716c from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Nucleoid-associated protein Rv3716c | | Authors: | Deka, G, Gopalan, A, Prabhavathi, M, Savithri, H.S, Raja, A, Murthy, M.R.N. | | Deposit date: | 2017-11-10 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical characterization of Rv3716c, a hypothetical protein from Mycobacterium tuberculosis
Biochem. Biophys. Res. Commun., 495, 2018
|
|
7A5I
 
 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
6Z8X
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
5ZB8
 
 | |
8OOK
 
 | |
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
1BL3
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
4IGK
 
 | |
6W62
 
 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XXC
 
 | | Ternary complex of 14-3-3 sigma (C38N), Estrogen Related Receptor gamma (DBD) phosphopeptide, and disulfide PPI stabilizer 4 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Estrogen Related Receptor gamma phosphopeptide, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2020-01-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
4IFI
 
 | |
6Z8V
 
 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
1BQN
 
 | | TYR 188 LEU HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQM
 
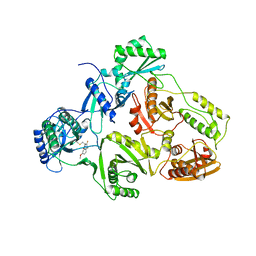 | | HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
3E5Q
 
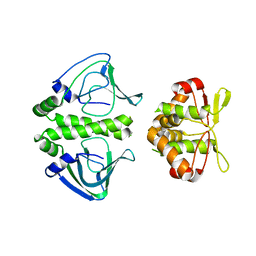 | | Unbound Oxidised CprK | | Descriptor: | Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
5HXY
 
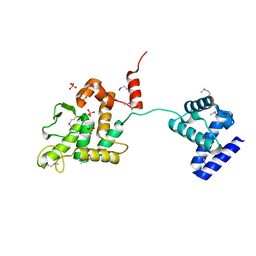 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
1H7U
 
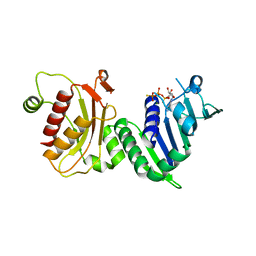 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
3E9W
 
 | |
1BHL
 
 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BI4
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-22 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1MM3
 
 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
2OUG
 
 | |
3E5X
 
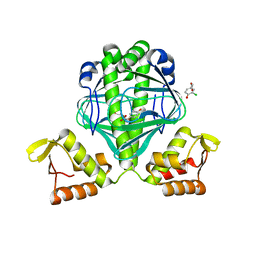 | | OCPA complexed CprK | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
1NFI
 
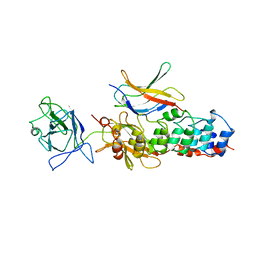 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
