1BCT
 
 | |
1BCU
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN AND PROFLAVIN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, PROFLAVIN | | Authors: | Conti, E, Rivetti, C, Wonacott, A, Brick, P. | | Deposit date: | 1998-05-02 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and spectrophotometric studies of the binding of proflavin to the S1 specificity pocket of human alpha-thrombin.
FEBS Lett., 425, 1998
|
|
1BCV
 
 | | SYNTHETIC PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS, NMR, 10 STRUCTURES | | Descriptor: | PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-03 | | Release date: | 1998-11-25 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1BCW
 
 | | RECOMBINANT RAT ANNEXIN V, T72A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCX
 
 | |
1BCY
 
 | | RECOMBINANT RAT ANNEXIN V, T72K MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCZ
 
 | | RECOMBINANT RAT ANNEXIN V, T72S MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
1BD1
 
 | | CRYSTALLOGRAPHIC STUDY OF ONE TURN OF G/C-RICH B-DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G)-3'), TRIETHYLAMMONIUM ION | | Authors: | Heinemann, U. | | Deposit date: | 1989-08-16 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic study of one turn of G/C-rich B-DNA.
J.Mol.Biol., 210, 1989
|
|
1BD2
 
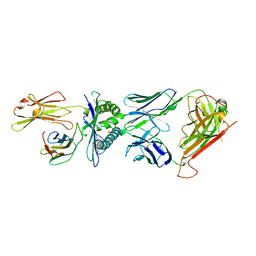 | | COMPLEX BETWEEN HUMAN T-CELL RECEPTOR B7, VIRAL PEPTIDE (TAX) AND MHC CLASS I MOLECULE HLA-A 0201 | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A 0201, T CELL RECEPTOR ALPHA, ... | | Authors: | Ding, Y.-H, Smith, K.J, Garboczi, D.N, Utz, U, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids.
Immunity, 8, 1998
|
|
1BD3
 
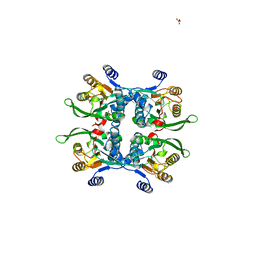 | | STRUCTURE OF THE APO URACIL PHOSPHORIBOSYLTRANSFERASE, 2 MUTANT C128V | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1BD4
 
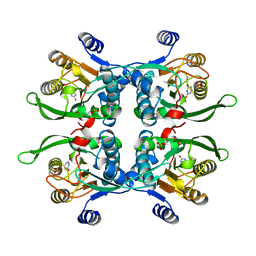 | | UPRT-URACIL COMPLEX | | Descriptor: | PHOSPHATE ION, URACIL, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-05-12 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1BD6
 
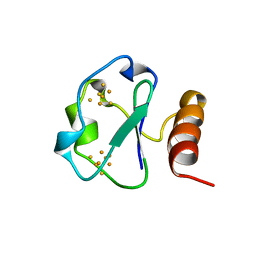 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BD7
 
 | | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Descriptor: | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Authors: | Wright, G, Basak, A.K, Mayr, E.-M, Glockshuber, R, Slingsby, C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circular permutation of betaB2-crystallin changes the hierarchy of domain assembly.
Protein Sci., 7, 1998
|
|
1BD8
 
 | | STRUCTURE OF CDK INHIBITOR P19INK4D | | Descriptor: | P19INK4D CDK4/6 INHIBITOR | | Authors: | Baumgartner, R, Fernandez-Catalan, C, Winoto, A, Huber, R, Engh, R, Holak, T.A. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human cyclin-dependent kinase inhibitor p19INK4d: comparison to known ankyrin-repeat-containing structures and implications for the dysfunction of tumor suppressor p16INK4a.
Structure, 6, 1998
|
|
1BD9
 
 | |
1BDA
 
 | |
1BDB
 
 | | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE FROM PSEUDOMONAS SP. LB400 | | Descriptor: | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Huelsmeyer, M, Hecht, H.-J, Niefind, K, Hofer, B, Timmis, K.N, Schomburg, D. | | Deposit date: | 1997-05-10 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
Protein Sci., 7, 1998
|
|
1BDC
 
 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1BDD
 
 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1BDE
 
 | | HELICAL STRUCTURE OF POLYPEPTIDES FROM THE C-TERMINAL HALF OF HIV-1 VPR, NMR, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1998-05-07 | | Release date: | 1998-12-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of peptides from HIV-1 Vpr protein that cause membrane permeabilization and growth arrest.
J.Pept.Sci., 4, 1998
|
|
1BDF
 
 | |
1BDG
 
 | |
1BDH
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Glasfeld, A, Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-07-25 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Residue Bound in the Minor Groove Does not Alter the Bending of a DNA Duplex
J.Am.Chem.Soc., 118, 1996
|
|
1BDI
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Glasfeld, A, Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-07-25 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Positively Charged Residue Bound in the Minor Groove Does not Alter the Bending of a DNA Duplex
J.Am.Chem.Soc., 118, 1996
|
|
