3D5O
 
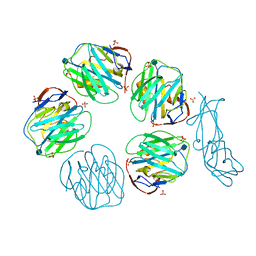 | | Structural recognition and functional activation of FcrR by innate pentraxins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low affinity immunoglobulin gamma Fc region receptor II-a, ... | | Authors: | Lu, J, Marnell, L.L, Marjon, K.D, Mold, C, Du Clos, T.W, Sun, P.D. | | Deposit date: | 2008-05-16 | | Release date: | 2008-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural recognition and functional activation of FcgammaR by innate pentraxins.
Nature, 456, 2008
|
|
4FP1
 
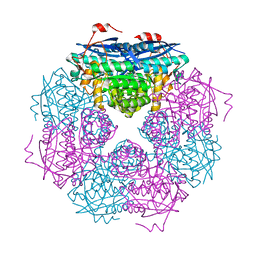 | | P. putida mandelate racemase co-crystallized with 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl) propionic acid | | Descriptor: | 3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propanoic acid, MAGNESIUM ION, Mandelate racemase | | Authors: | Lietzan, A.D, St.Maurice, M. | | Deposit date: | 2012-06-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Potent inhibition of mandelate racemase by a fluorinated substrate-product analogue with a novel binding mode.
Biochemistry, 53, 2014
|
|
3D5X
 
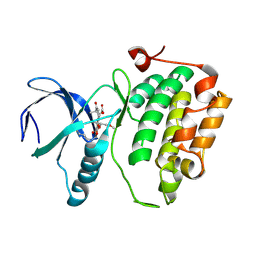 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with wortmannin. | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Polo-like kinase 1 | | Authors: | Elling, R.A, Fucini, R.V, Romanowski, M.J. | | Deposit date: | 2008-05-17 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the wild-type and activated catalytic domains of Brachydanio rerio Polo-like kinase 1 (Plk1): changes in the active-site conformation and interactions with ligands.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4FMO
 
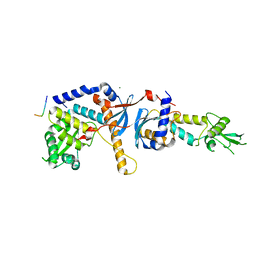 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3D68
 
 | |
3D72
 
 | |
3D3Y
 
 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-13 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
3D7K
 
 | | Crystal structure of benzaldehyde lyase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S. | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Probing the active center of benzaldehyde lyase with substitutions and the pseudosubstrate analogue benzoylphosphonic acid methyl ester
Biochemistry, 47, 2008
|
|
4FQA
 
 | |
3VNP
 
 | | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus | | Descriptor: | ACETIC ACID, Hypothetical conserved protein, MAGNESIUM ION | | Authors: | Karthe, P.P, Kumarevel, T.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus
To be Published
|
|
4FNI
 
 | |
4FNS
 
 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E from Geobacillus stearothermophilus in complex with 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-galactosidase AgaA, ... | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
3D59
 
 | |
3CEZ
 
 | | Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei | | Descriptor: | ACETIC ACID, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Staker, B, Napuli, A, Nakazawa, S.H, Castaneda, L, Alkafeef, S, Vanvoorhis, W, Stewart, L, Myler, P, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine-R-sulfoxide reductase from Burkholderia pseudomallei.
To be Published
|
|
4FQU
 
 | |
3D6J
 
 | | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative haloacid dehalogenase-like hydrolase | | Authors: | Chang, C, Wu, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis
To be Published
|
|
4FP8
 
 | |
3D6N
 
 | | Crystal Structure of Aquifex Dihydroorotase Activated by Aspartate Transcarbamoylase | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, Dihydroorotase, ... | | Authors: | Edwards, B.F.P. | | Deposit date: | 2008-05-20 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.
Biochemistry, 48, 2009
|
|
4FR0
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with SAM | | Descriptor: | Arsenic methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
4FHI
 
 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, SRC-1, Vitamin Nuclear Receptor | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
3CFE
 
 | | Crystal structure of purple-fluorescent antibody EP2-25C10 | | Descriptor: | GLYCEROL, PURPLE-FLUORESCENT ANTIBODY EP2-25C10-IGG2B HEAVY CHAIN, PURPLE-FLUORESCENT ANTIBODY EP2-25C10-KAPPA LIGHT CHAIN, ... | | Authors: | Debler, E.W, Heine, A, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3D75
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, at pH 5.5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
4FHP
 
 | | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity - CaUTP bound | | Descriptor: | CALCIUM ION, GLYCEROL, Poly(A) RNA polymerase protein cid1, ... | | Authors: | Lunde, B.M, Magler, I, Meinhart, A. | | Deposit date: | 2012-06-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity.
Nucleic Acids Res., 40, 2012
|
|
3CFP
 
 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
3CGE
 
 | |
