4DVY
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
2VS5
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2VS3
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
6QXL
 
 | | Crystal Structure of Pyruvate Kinase II (PykA) from Pseudomonas aeruginosa in complex with sodium malonate, magnesium and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Abdelhamid, Y, Brear, P, Welch, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA fromPseudomonas aeruginosa.
J.Biol.Chem., 294, 2019
|
|
2VS4
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
4DSY
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
4FXL
 
 | | Crystal structure of the D76N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Bellotti, V, Pepys, M.B, Stoppini, M, Bolognesi, M. | | Deposit date: | 2012-07-03 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hereditary systemic amyloidosis due to Asp76Asn variant beta-2-microglobulin.
N.Engl.J.Med., 366, 2012
|
|
1NOI
 
 | |
4DT2
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC27209 | | Descriptor: | (2,2-dimethyl-2,3-dihydro-1-benzofuran-7-yl)methanol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
4F3R
 
 | | Structure of phosphopantetheine adenylyltransferase (CBU_0288) from Coxiella burnetii | | Descriptor: | CALCIUM ION, Phosphopantetheine adenylyltransferase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
1T2N
 
 | |
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
1T4M
 
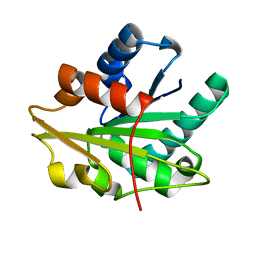 | |
1CXY
 
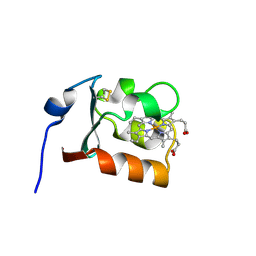 | | STRUCTURE AND CHARACTERIZATION OF ECTOTHIORHODOSPIRA VACUOLATA CYTOCHROME B558, A PROKARYOTIC HOMOLOGUE OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kostanjevecki, V, Leys, D, Van Driessche, G, Meyer, T.E, Cusanovich, M.A, Fischer, U, Guisez, Y, Van Beeumen, J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and characterization of Ectothiorhodospira vacuolata cytochrome b(558), a prokaryotic homologue of cytochrome b(5).
J.Biol.Chem., 274, 1999
|
|
1DLJ
 
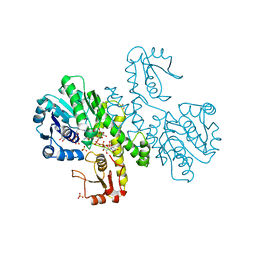 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
1H8U
 
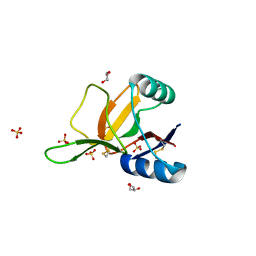 | | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A: An Atypical Lectin with a Paradigm Shift in Specificity | | Descriptor: | EOSINOPHIL GRANULE MAJOR BASIC PROTEIN 1, GLYCEROL, SULFATE ION | | Authors: | Swaminathan, G.J, Weaver, A.J, Loegering, D.A, Checkel, J.L, Leonidas, D.D, Gleich, G.J, Acharya, K.R. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A. An Atypical Lectin with a Paradigm Shift in Specificity
J.Biol.Chem., 276, 2001
|
|
3LEA
 
 | |
3IOV
 
 | |
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IO6
 
 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IOU
 
 | |
2J4T
 
 | | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein | | Descriptor: | ANGIOGENIN-4 | | Authors: | Crabtree, B, Holloway, D.E, Baker, M.D, Acharya, K.R, Subramanian, V. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein
Biochemistry, 46, 2007
|
|
3LE9
 
 | |
