4Q2N
 
 | |
7MIQ
 
 | | Crystal structure of a Glutathione S-transferase class Gtt2 of Vibrio parahaemolyticus (VpGSTT2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutathione S-transferase, SULFATE ION | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A.A, Garcia-Orozco, K.D, Sotelo-Mundo, R.R. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Novel Glutathione S -Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen.
Toxins, 13, 2021
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
4MZJ
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[801,805], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, Myosin-A | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
6HW1
 
 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
4MZQ
 
 | |
4IBM
 
 | | Crystal structure of insulin receptor kinase domain in complex with an inhibitor Irfin-1 | | Descriptor: | 5-(2-phenylpyrazolo[1,5-a]pyridin-3-yl)-3H-pyrazolo[3,4-c]pyridazin-3-one, Insulin receptor | | Authors: | Wu, J, Anastassiadis, T, Duong-Ly, K.C, Peterson, J.R. | | Deposit date: | 2012-12-08 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor.
J.Biol.Chem., 288, 2013
|
|
4P76
 
 | | Cellular response to a crystal-forming protein | | Descriptor: | Photoconvertible fluorescent protein, SODIUM ION | | Authors: | Tsutsui, H, Jinno, Y, Shoda, K, Tomita, A, Matsuda, M, Yamashita, E, Katayama, H, Nakagawa, A, Miyawaki, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A diffraction-quality protein crystal processed as an autophagic cargo
Mol.Cell, 58, 2015
|
|
5EU5
 
 | | HLA Class I antigen | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Bianchi, V, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A Molecular Switch Abrogates Glycoprotein 100 (gp100) T-cell Receptor (TCR) Targeting of a Human Melanoma Antigen.
J.Biol.Chem., 291, 2016
|
|
4Z7X
 
 | | MdbA protein, a thiol-disulfide oxidoreductase from Actinomyces oris. | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, MdbA | | Authors: | OSIPIUK, J, Reardon-Robinson, M.E, Ton-That, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris.
J.Biol.Chem., 290, 2015
|
|
4MZK
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[807,811], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, pGly[807,811], a stapled myoA tail peptide | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
3PLD
 
 | | Endothiapepsin in complex with a fragment | | Descriptor: | 4-chlorobenzyl carbamimidothioate, Endothiapepsin, GLYCEROL | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
4MYO
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NB3
 
 | | Crystal structure of RPA70N in complex with a 3,4 dichlorophenylalanine ATRIP derived peptide | | Descriptor: | 3,4 dichlorophenylalanine ATRIP derived peptide, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Souza-Fagundes, E.M, Luzwik, J.W, Cortez, D, Olejniczak, O.T, Waterson, A.G, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
J.Med.Chem., 57, 2014
|
|
4R8W
 
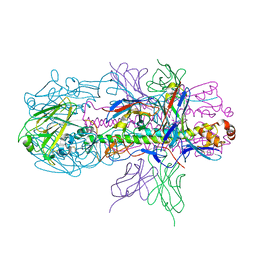 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | Authors: | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
3PGI
 
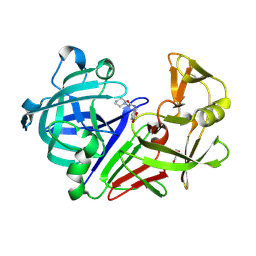 | | Endothiapepsin in complex with a fragment | | Descriptor: | Endothiapepsin, GLYCEROL, N-(1,3-benzodioxol-5-yl)-2-(piperidin-1-yl)acetamide | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
5ORL
 
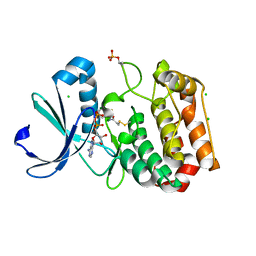 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, Vrzal, L, Birchall, K, Arnold, L.H, Mpamhanga, C, Coombs, P.J, Burgess, S.G, Richards, M.W, Winter, A, Veverka, V, von Delft, F, Merritt, A, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
1YWT
 
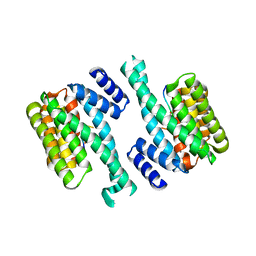 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
7LWG
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12694 | | Descriptor: | 5-{(5S)-1-[2-({3-chloro-6-[(2S)-2,4-dimethylpiperazin-1-yl]-2-fluoropyridin-4-yl}amino)-2-oxoethyl]-4-oxo-4,6,7,8-tetrahydro-1H-dipyrrolo[1,2-a:2',3'-d]pyrimidin-3-yl}-3,4-difluoro-2-hydroxybenzamide, B-cell lymphoma 6 protein, GLYCEROL, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
4BHN
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
1M2T
 
 | | Mistletoe Lectin I from Viscum album in Complex with Adenine Monophosphate. Crystal Structure at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Krauspenhaar, R, Rypniewski, W, Kalkura, N, Moore, K, DeLucas, L, Stoeva, S, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallisation under microgravity of mistletoe lectin I from Viscum album with adenine monophosphate and the crystal structure at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2XNE
 
 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
6CO6
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2XNG
 
 | | Structure of Aurora-A bound to a selective imidazopyrazine inhibitor | | Descriptor: | N-(3-{3-chloro-8-[(4-morpholin-4-ylphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzyl)methanesulfonamide, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
5O0S
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
