2OF6
 
 | | Structure of immature West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, Y, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of immature west nile virus.
J.Virol., 81, 2007
|
|
3CK2
 
 | | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Conserved uncharacterized protein (predicted phosphoesterase COG0622), MANGANESE (II) ION, ... | | Authors: | Nocek, B, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4.
To be Published
|
|
4JPF
 
 | | Structure of wild type Pseudomonas aeruginosa FabF (KASII) in Complex with ligand | | Descriptor: | 3-(benzoylamino)-2-hydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, MAGNESIUM ION, ... | | Authors: | Baum, B, Brenk, R, Jaenicke, E. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structures of Pseudomonas aeruginosa beta-ketoacyl-(acyl-carrier-protein) synthase II (FabF) and a C164Q mutant provide templates for antibacterial drug discovery and identify a buried potassium ion and a ligand-binding site that is an artefact of the crystal form.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
6W43
 
 | | APE1 AP-endonuclease product complex R237C | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
1R7J
 
 | | Crystal structure of the DNA-binding protein Sso10a from Sulfolobus solfataricus | | Descriptor: | Conserved hypothetical protein Sso10a | | Authors: | Chen, L, Chen, L.R, Zhou, X.E, Wang, Y, Kahsai, M.A, Clark, A.T, Edmondson, S.P, Liu, Z.-J, Rose, J.P, Wang, B.C, Shriver, J.W, Meehan, E.J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod.
J.Mol.Biol., 341, 2004
|
|
5NHT
 
 | | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-03-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | human 199.54-16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
1RC8
 
 | | T4 Polynucleotide Kinase bound to 5'-GTCAC-3' ssDNA | | Descriptor: | 5'-D(*GP*TP*CP*AP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-11-03 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RYB
 
 | |
4YHZ
 
 | | Crystal structure of 304M3-B Fab in complex with H3K4me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4J67
 
 | | Crystal structure of Ribonuclease A soaked in 50% 1,6-Hexanediol: One of twelve in MSCS set | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
8FXF
 
 | | Crystal structure of the coiled-coil domain of TRIM56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase TRIM56 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.
Comput Struct Biotechnol J, 21, 2023
|
|
2O22
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-2, N-[(4-{[1,1-dimethyl-2-(phenylthio)ethyl]amino}-3-nitrophenyl)sulfonyl]-4-(4,4-dimethylpiperidin-1-yl)benzamide | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O2M
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6Y0M
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N mutant | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
1RYN
 
 | |
7D35
 
 | | Human LC8 bound to ebola virus VP35(67-76) | | Descriptor: | Dynein light chain 1, cytoplasmic, Peptide from Polymerase cofactor VP35 | | Authors: | Ku, B, Lim, D. | | Deposit date: | 2020-09-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of human LC8 bound to a peptide from Ebola virus VP35.
J.Microbiol, 59, 2021
|
|
2O64
 
 | | Crystal structure of Pim1 with Quercetagetin | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O1Y
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
6RBM
 
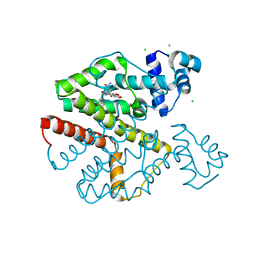 | | TETR(D) E147A MUTANT IN COMPLEX WITH MINOCYCLINE AND MAGNESIUM | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hinrichs, W, Palm, G.J, Berndt, L, Girbardt, B. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
1S8G
 
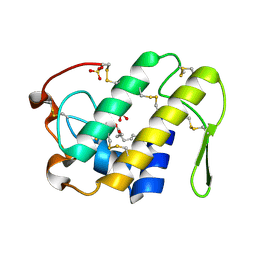 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form | | Descriptor: | GLYCEROL, LAURIC ACID, Phospholipase A2 homolog, ... | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
4JG1
 
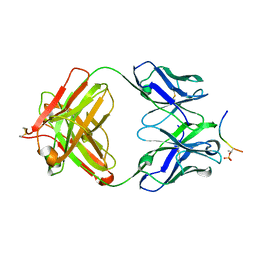 | | Structure of phosphoserine/threonine (pSTAb) scaffold bound to pThr peptide | | Descriptor: | Fab heavy chain, Fab light chain, PROPANOIC ACID, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
2O3P
 
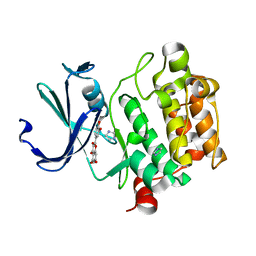 | | Crystal structure of Pim1 with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-01 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
1S4A
 
 | |
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
