6RXV
 
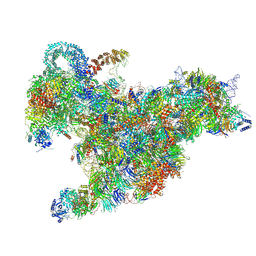 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
2QF2
 
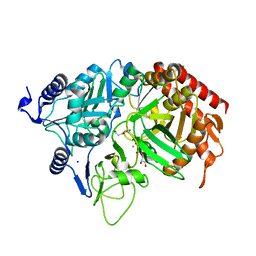 | | Rat cytosolic PEPCK in complex with oxaloacetic acid and GDP. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, OXALOACETATE ION, ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
9BDN
 
 | |
9BDP
 
 | |
2QF1
 
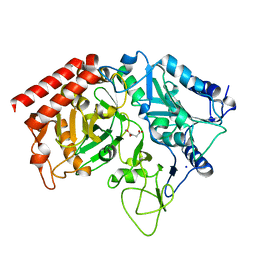 | | Rat cytosolic PEPCK in complex with oxaloacetic acid. | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Phosphoenolpyruvate carboxykinase, ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
6RXY
 
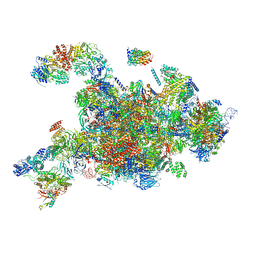 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
2QEW
 
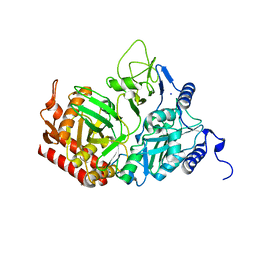 | | Rat cytosolic PEPCK, in complex with manganese ion. | | Descriptor: | MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, cytosolic [GTP], ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
8H7G
 
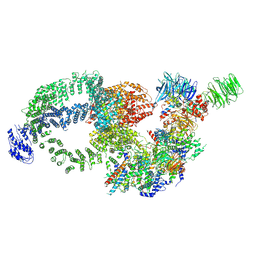 | | Cryo-EM structure of the human SAGA complex | | Descriptor: | Ataxin-7, STAGA complex 65 subunit gamma, Splicing factor 3B subunit 3, ... | | Authors: | Huang, J, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human SAGA transcriptional coactivator complex.
Cell Discov, 8, 2022
|
|
7YPB
 
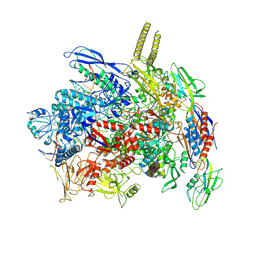 | |
6RXZ
 
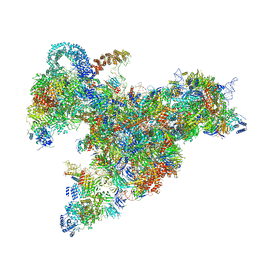 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXU
 
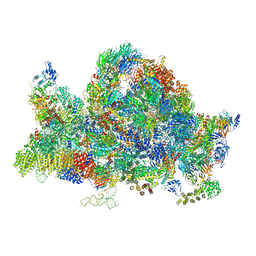 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXT
 
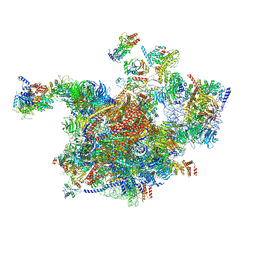 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
7VPX
 
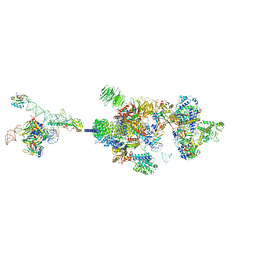 | | The cryo-EM structure of the human pre-A complex | | Descriptor: | 5SS, DnaJ homolog subfamily C member 8, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-10-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7QGH
 
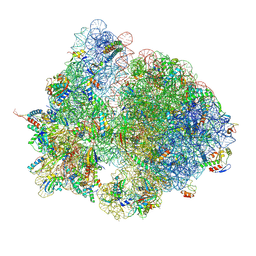 | | Structure of the E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
4E3S
 
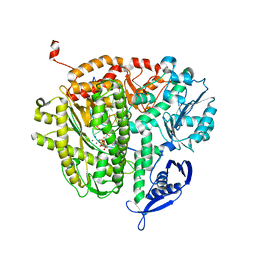 | | RB69 DNA Polymerase Ternary Complex with dQTP Opposite dT | | Descriptor: | 3-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-7-methyl-3H-imidazo[4,5-b]pyridine, CALCIUM ION, DNA polymerase, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-03-10 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing minor groove hydrogen bonding interactions between RB69 DNA polymerase and DNA.
Biochemistry, 51, 2012
|
|
4DH1
 
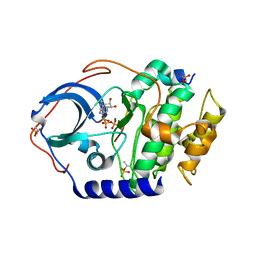 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH8
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3BRI
 
 | | Crystal Structure of apo-LC8 | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E, Chiodo, M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interplay of Ligand Binding and Quaternary Structure in the Diverse Interactions of Dynein Light Chain LC8.
J.Mol.Biol., 384, 2008
|
|
6KJ6
 
 | | cryo-EM structure of Escherichia coli Crl transcription activation complex | | Descriptor: | DNA (51-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Xu, J, Zhang, Y. | | Deposit date: | 2019-07-21 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.
Elife, 8, 2019
|
|
9ICW
 
 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity
Biochemistry, 35, 1996
|
|
9ICM
 
 | | DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DOUBLE STRANDED DNA (NO 5'-PHOSPHATE) | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*TP*GP*T)-3'), PROTEIN (DNA POLYMERASE BETA (E.C.2.7.7.7)), ... | | Authors: | Pelletier, H, Sawaya, M.R. | | Deposit date: | 1995-12-16 | | Release date: | 1996-11-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity
Biochemistry, 35, 1996
|
|
7KTS
 
 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6NMI
 
 | | Cryo-EM structure of the human TFIIH core complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription and DNA repair factor IIH helicase subunit XPD, ... | | Authors: | Greber, B.J, Toso, D, Fang, J, Nogales, E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The complete structure of the human TFIIH core complex.
Elife, 8, 2019
|
|
6DGP
 
 | |
4W9S
 
 | | 2-(4-(1H-tetrazol-5-yl)phenyl)-5-hydroxypyrimidin-4(3H)-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 5-hydroxy-2-[4-(1H-tetrazol-5-yl)phenyl]pyrimidin-4(3H)-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phenyl Substituted 4-Hydroxypyridazin-3(2H)-ones and 5-Hydroxypyrimidin-4(3H)-ones: Inhibitors of Influenza A Endonuclease.
J.Med.Chem., 57, 2014
|
|
