2XO5
 
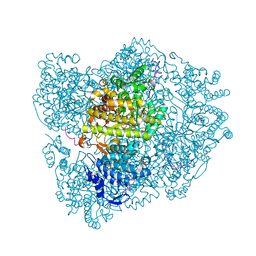 | | RIBONUCLEOTIDE REDUCTASE Y731NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
1Z9H
 
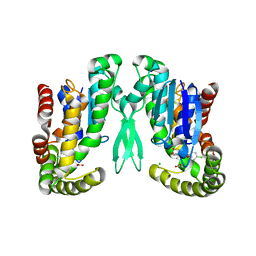 | | Microsomal prostaglandin E synthase type-2 | | Descriptor: | ACETATE ION, CHLORIDE ION, INDOMETHACIN, ... | | Authors: | Yamada, T, Komoto, J, Watanabe, K, Ohmiya, Y, Takusagawa, F. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Possible Catalytic Mechanism of Microsomal Prostaglandin E Synthase Type 2 (mPGES-2).
J.Mol.Biol., 348, 2005
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
1ZKX
 
 | | Crystal structure of Glu158Ala/Thr159Ala/Asn160Ala- a triple mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
5T36
 
 | | Crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 4-chloro-2-[({(1S,2S)-2-[(2,2-dimethylpropanoyl)amino]cyclopentyl}methyl)amino]benzoic acid, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3GGQ
 
 | | Dimerization of Hepatitis E Virus Capsid Protein E2s Domain is Essential for Virus-Host Interaction | | Descriptor: | BROMIDE ION, Capsid protein | | Authors: | Li, S.W, Tang, X.H, Seetharaman, J, Yang, C.Y, Gu, Y, Zhang, J, Du, H.L, Shih, J.W.K, Hew, C.L, Sivaraman, J, Xia, N.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction
Plos Pathog., 5, 2009
|
|
5T37
 
 | | crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 2-chloro-5-{[(2,2-dimethylpropanoyl)amino]methyl}-N-(1H-imidazol-2-yl)benzamide, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6CJ5
 
 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-3-yl)imidazo[1,2-a]pyridine-8-carboxamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CK6
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 6'-[(6-aminopyrimidin-4-yl)amino]-8'-methyl-2'H-spiro[cyclohexane-1,3'-imidazo[1,5-a]pyridine]-1',5'-dione, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CKI
 
 | | Co-crystal structure of MNK2 in Complex With Inhibitor | | Descriptor: | 3,3-dimethyl-6-[(pyrimidin-4-yl)amino]-2,3-dihydroimidazo[1,5-a]pyridine-1,5-dione, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
5N2R
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with PSB36 at 2.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-butyl-3-[(~{E})-3-oxidanylprop-1-enyl]-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, Adenosine receptor A2a,Soluble cytochrome b562,ADENOSINE RECEPTOR A2A SOLUBLE CYTOCHROME B562 ADENOSINE RECEPTOR A2A,Adenosine receptor A2a, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
7KQB
 
 | | SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 5A6 heavy chain, Fab 5A6 light chain, ... | | Authors: | Asarnow, D, Charles, C, Cheng, Y. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
8JXQ
 
 | | Cryo-EM structure of bilirubin ditaurate (BDT) bound human ABC transporter ABCC2 | | Descriptor: | 2-[3-[5-[(E)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-3-[3-oxidanylidene-3-(2-sulfoethylamino)propyl]-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoylamino]ethanesulfonic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL | | Authors: | Mao, Y.X, Chen, Z.P, Wang, L, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.
Nat Commun, 15, 2024
|
|
7KRF
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-(3-(2-cyanovinyl)-5-fluorophenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ710) | | Descriptor: | 4-{3-[(E)-2-cyanoethenyl]-5-fluorophenoxy}-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
1LTA
 
 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | Authors: | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
9ARG
 
 | |
6BAO
 
 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
3G0E
 
 | | KIT kinase domain in complex with sunitinib | | Descriptor: | Mast/stem cell growth factor receptor, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Gajiwala, K.S, Wu, J.C, Lunney, E.A, Gemetri, G.D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GME
 
 | |
7L1X
 
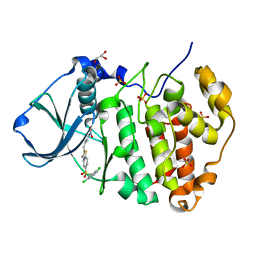 | | Structure of human CK2 alpha kinase (catalytic subunit) with the inhibitor 108600. | | Descriptor: | (2~{Z})-6-[[2,6-bis(chloranyl)phenyl]methylsulfonyl]-2-[[4-oxidanyl-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methylidene]-4~{H}-1,4-benzothiazin-3-one, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous CK2/TNIK/DYRK1 inhibition by 108600 suppresses triple negative breast cancer stem cells and chemotherapy-resistant disease.
Nat Commun, 12, 2021
|
|
6CJH
 
 | | Co-crystal structure of MNK2 in complex with an inhibitor | | Descriptor: | 3-phenyl-5-(pyridin-4-yl)-1H-indazole, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
2XO4
 
 | | RIBONUCLEOTIDE REDUCTASE Y730NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
3MV0
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (R599A) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
5TL9
 
 | | crystal structure of mPGES-1 bound to inhibitor | | Descriptor: | 2-{2-[(1S,2S)-2-{[1-(8-methylquinolin-2-yl)piperidine-4-carbonyl]amino}cyclopentyl]ethyl}benzoic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Partridge, K, Fisher, M. | | Deposit date: | 2016-10-10 | | Release date: | 2017-03-01 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and characterization of [(cyclopentyl)ethyl]benzoic acid inhibitors of microsomal prostaglandin E synthase-1.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
