3FWC
 
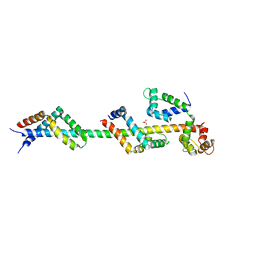 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1, ... | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
3QR6
 
 | |
3FXU
 
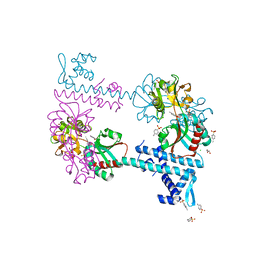 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3FZ1
 
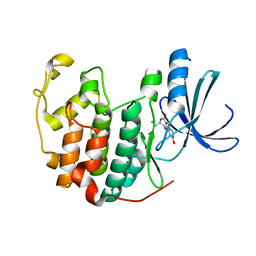 | | Crystal structure of a benzthiophene inhibitor bound to human Cyclin-dependent Kinase-2 (CDK-2) | | Descriptor: | (3R)-3-(aminomethyl)-9-methoxy-1,2,3,4-tetrahydro-5H-[1]benzothieno[3,2-e][1,4]diazepin-5-one, Cell division protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2009-01-23 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Benzothiophene inhibitors of MK2. Part 2: improvements in kinase selectivity and cell potency.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FZ7
 
 | |
3G8A
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoHcy in space group P61 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3QW9
 
 | | Crystal structure of betaglycan ZP-C domain | | Descriptor: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lin, S.J, Jardetzky, T.S. | | Deposit date: | 2011-02-27 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QWQ
 
 | |
3GAG
 
 | |
3QUS
 
 | | Crystal Structure of N10-Formyltetrahydrofolate Synthetase with ATPgS | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Formate--tetrahydrofolate ligase, MALEIC ACID, ... | | Authors: | Celeste, L.R, Chai, G, Bielak, M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2011-02-24 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Strucutre of N10-Formyltetrahydrofolate Synthetase with ATPgS
To be Published
|
|
3G0R
 
 | | Complex of Mth0212 and an 8bp dsDNA with distorted ends | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G1B
 
 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | Descriptor: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G1K
 
 | |
3GB8
 
 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
3GBI
 
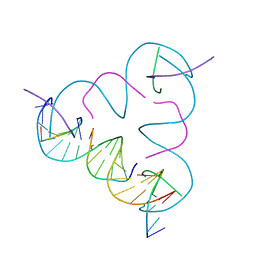 | | The Rational Design and Structural Analysis of a Self-Assembled Three-Dimensional DNA Crystal | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Birktoft, J.J, Zheng, J, Seeman, N.C. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal.
Nature, 461, 2009
|
|
3QZ4
 
 | |
3QY3
 
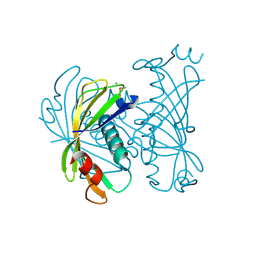 | | PA2801 protein, a putative Thioesterase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Thioesterase | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3G40
 
 | |
3QQB
 
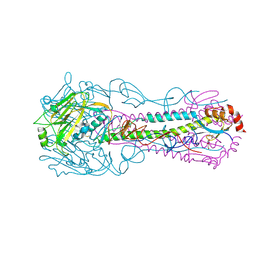 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, neutral pH form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
3QSR
 
 | |
3QR5
 
 | |
3GCV
 
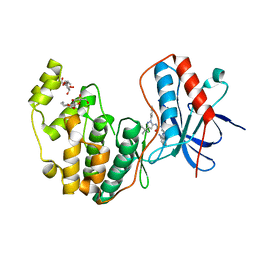 | | Human P38 MAP Kinase in Complex with RL62 | | Descriptor: | 1-{3-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
3G6I
 
 | |
3QTH
 
 | |
3GD7
 
 | | Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP) | | Descriptor: | Fusion complex of Cystic fibrosis transmembrane conductance regulator, residues 1193-1427 and Maltose/maltodextrin import ATP-binding protein malK, residues 219-371, ... | | Authors: | Atwell, S, Antonysamy, S, Conners, K, Emtage, S, Gheyi, T, Lewis, H.A, Lu, F, Sauder, J.M, Wasserman, S.R, Zhao, X. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP)
To be Published
|
|
