7UCC
 
 | | Transcription factor FosB/JunD bZIP domain in the reduced form | | Descriptor: | CHLORIDE ION, ETHANOL, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
6SDG
 
 | |
4GVP
 
 | |
6FI9
 
 | |
8YQV
 
 | | African swine fever virus RNA Polymerase core | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Regulation of early transcription and late transcript processing enzymes packaging for African swine fever virus from endogenous vRNAP-M1249L supercomplex structures
To Be Published
|
|
2WEV
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-B3L-MEA, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncation and Optimisation of Peptide Inhibitors of Cyclin-Dependent Kinase 2-Cyclin a Through Structure-Guided Design.
Chemmedchem, 4, 2009
|
|
2WFY
 
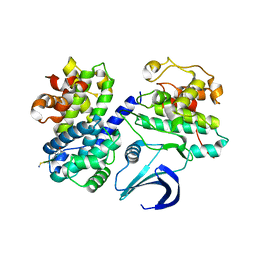 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-B3L-PHE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Truncation and Optimisation of Peptide Inhibitors of Cyclin-Dependent Kinase 2-Cyclin a Through Structure-Guided Design.
Chemmedchem, 4, 2009
|
|
1E3F
 
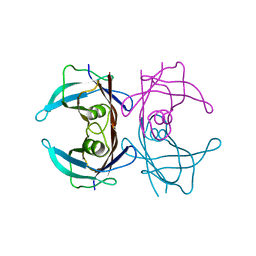 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-06-14 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2WHB
 
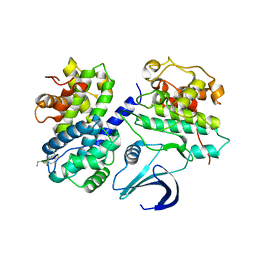 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-L3O-PFF, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-05-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Truncation and optimisation of peptide inhibitors of cyclin-dependent kinase 2-cyclin a through structure-guided design.
Chemmedchem, 4, 2009
|
|
1E4H
 
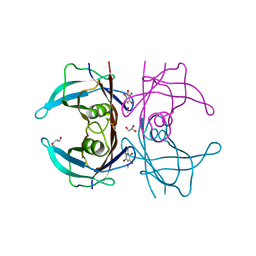 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | GLYCEROL, PENTABROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-04 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8YQY
 
 | | ASFV RNA polymerase-M1249L complex complete | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Regulation of early transcription and late transcript processing enzymes packaging for African swine fever virus from endogenous vRNAP-M1249L supercomplex structures
To Be Published
|
|
8YQU
 
 | | African swine fever virus RNA Polymerase-M1249L complex1 | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Regulation of early transcription and late transcript processing enzymes packaging for African swine fever virus from endogenous vRNAP-M1249L supercomplex structures
To Be Published
|
|
8YQT
 
 | | African swine fever virus RNA Polymerase-M1249L complex2 | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Regulation of early transcription and late transcript processing enzymes packaging for African swine fever virus from endogenous vRNAP-M1249L supercomplex structures
To Be Published
|
|
8YQW
 
 | | ASFV RNA polymerase-M1249L complex3 | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | ASFV RNA polymerase-M1249L complex3
To Be Published
|
|
4IF4
 
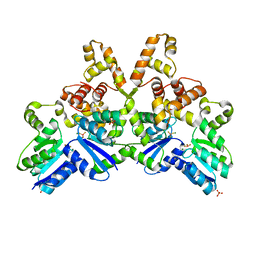 | |
1HMF
 
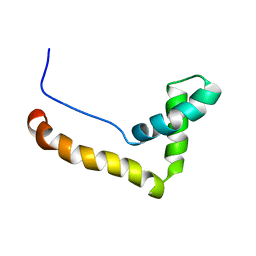 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
7O9M
 
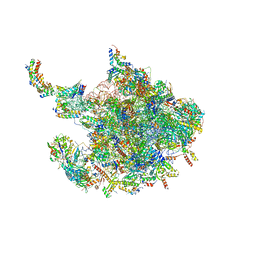 | | Human mitochondrial ribosome large subunit assembly intermediate with MTERF4-NSUN4, MRM2, MTG1 and the MALSU module | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Hallberg, B.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for late maturation steps of the human mitoribosomal large subunit.
Nat Commun, 12, 2021
|
|
1P4X
 
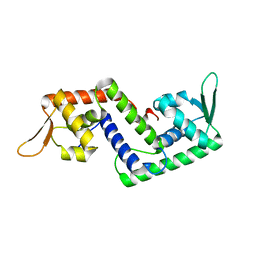 | | Crystal structure of SarS protein from Staphylococcus Aureus | | Descriptor: | staphylococcal accessory regulator A homologue | | Authors: | Li, R, Manna, A.C, Dai, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SarS protein from Staphylococcus aureus
J.BACTERIOL., 185, 2003
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
6G16
 
 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
4B3A
 
 | | Tetracycline repressor class D mutant H100A in complex with tetracycline | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | Authors: | Eltschkner, S, Schindler, S, Palm, G.J, Schneider, J, Hinrichs, W. | | Deposit date: | 2012-07-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
5DCL
 
 | | Structure of a lantibiotic response regulator: N terminal domain of the nisin resistance regulator NsrR | | Descriptor: | 1,2-ETHANEDIOL, PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
5DCM
 
 | | Structure of a lantibiotic response regulator: C-terminal domain of the nisin resistance regulator NsrR | | Descriptor: | PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H.J. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
3KP3
 
 | | Staphylococcus epidermidis in complex with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Transcriptional regulator TcaR | | Authors: | Chang, Y.M, Chen, C.K, Wang, A.H. | | Deposit date: | 2009-11-15 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
