8GBJ
 
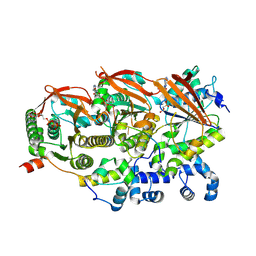 | | Cryo-EM structure of a human BCDX2/ssDNA complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*C)-3'), DNA repair protein RAD51 homolog 2, DNA repair protein RAD51 homolog 3, ... | | Authors: | Jia, L, Wasmuth, E.V, Ruben, E.A, Sung, P, Rawal, Y, Greene, E.C, Meir, A, Olsen, S.K. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into BCDX2 complex function in homologous recombination.
Nature, 619, 2023
|
|
8J9G
 
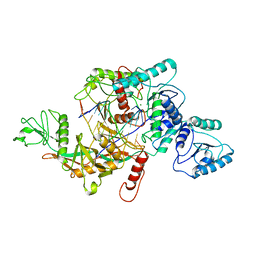 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
5J9R
 
 | |
5J84
 
 | |
8J8H
 
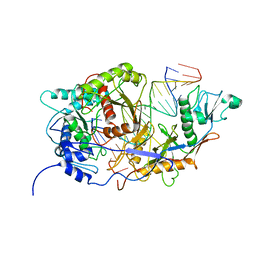 | | SPARTA monomer bound with guide-target, state 2 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
6N2E
 
 | |
5JFT
 
 | | Zebra Fish Caspase-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACE-ASP-GLU-VAL-ASK, ... | | Authors: | Tucker, M.B, MacKenzie, S.H, Maciag, J.J, Dirscherl, H, Swartz, P.D, Yoder, J.A, Hamilton, P.T, Clark, A.C. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish.
Protein Sci., 25, 2016
|
|
4XZ2
 
 | | Human platelet phosphofructokinase in an R-state in complex with ADP and F6P, crystal form I | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kloos, M, Strater, N. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation.
Biochem.J., 469, 2015
|
|
4XIM
 
 | |
8AMU
 
 | | RepB pMV158 OBD domain bound to DDR region | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Amodio, J, Machon, C, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
6POE
 
 | | Structure of ACLY in complex with CoA | | Descriptor: | ATP-citrate synthase, COENZYME A | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VMF
 
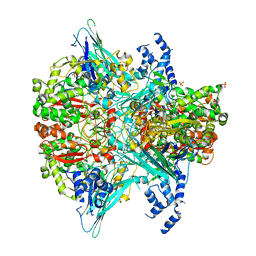 | | Crystal structure of the Y766F mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SULFATE ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
4YW6
 
 | | Structural Insight into Divalent Galactoside Binding to Pseudomonas aeruginosa lectin LecA | | Descriptor: | CALCIUM ION, N-[(2S)-6-amino-1-oxo-1-(pyrrolidin-1-yl)hexan-2-yl]-4-(beta-D-galactopyranosyloxy)benzamide, PA-I galactophilic lectin | | Authors: | Visini, R, Jin, X, Michaud, G, Bergmann, M, Gillon, E, Imberty, A, Stocker, A, Darbre, T, Pieters, R, Reymond, J.-L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insight into Multivalent Galactoside Binding to Pseudomonas aeruginosa Lectin LecA.
Acs Chem.Biol., 10, 2015
|
|
8IYQ
 
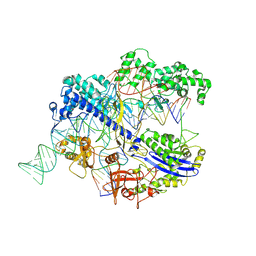 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
6PK7
 
 | |
6MWQ
 
 | |
6POF
 
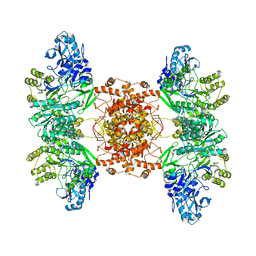 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4YSV
 
 | |
8KA5
 
 | |
8KA3
 
 | |
6PCF
 
 | | Human Coa6: W59C mutant protein | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog | | Authors: | Maher, M.J, Maghool, S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6.
Life Sci Alliance, 2, 2019
|
|
8OHA
 
 | | Crystal structure of Leptospira interrogans GAPDH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Navas-Yuste, S, de la Paz, K, Querol-Garcia, J, Gomez-Quevedo, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Front Immunol, 14, 2023
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7XPB
 
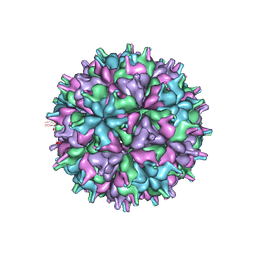 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5 | | Descriptor: | Capsid protein alpha | | Authors: | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | Deposit date: | 2022-05-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
6MYX
 
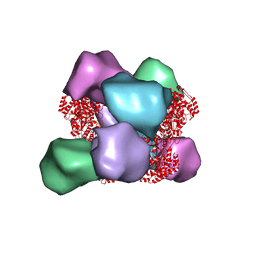 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
