7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
1FQV
 
 | | Insights into scf ubiquitin ligases from the structure of the skp1-skp2 complex | | Descriptor: | SKP1, SKP2 | | Authors: | Schulman, B.A, Carrano, A.C, Jeffrey, P.D, Bowen, Z, Kinnucan, E.R, Finnin, M.S, Elledge, S.J, Harper, J.W, Pagano, M, Pavletich, N.P. | | Deposit date: | 2000-09-06 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex.
Nature, 408, 2000
|
|
5XMW
 
 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1FFJ
 
 | | NMR STRUCTURE OF CARDIOTOXIN IN DPC-MICELLE | | Descriptor: | CYTOTOXIN 2 | | Authors: | Dubovskii, P.V, Dementieva, D.V, Bocharov, E.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Membrane binding motif of the P-type cardiotoxin.
J.Mol.Biol., 305, 2001
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
6Y15
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E_Q126K | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6Y1B
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
1FB5
 
 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
6Y8J
 
 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y5M
 
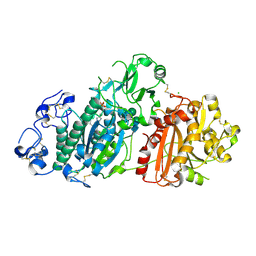 | | Crystal structure of mouse Autotaxin in complex with compound 1a | | Descriptor: | (~{E})-3-[4-chloranyl-2-[(5-methyl-1,2,3,4-tetrazol-2-yl)methyl]phenyl]-1-[(2~{R})-4-[(4-fluorophenyl)methyl]-2-methyl-piperazin-1-yl]prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Development of autotaxin inhibitors: A series of tetrazole cinnamides.
Bioorg.Med.Chem.Lett., 31, 2021
|
|
8QC4
 
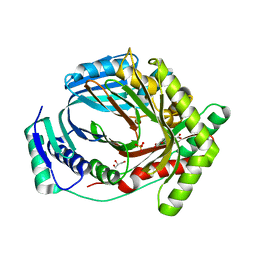 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-carboxyphenyl)furan-2-carboxylic acid | | Descriptor: | 5-(3-carboxyphenyl)furan-2-carboxylic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Structural Study of a New MbtI-Inhibitor Complex: Towards an Optimized Model for Structure-Based Drug Discovery.
Pharmaceuticals, 16, 2023
|
|
5CLB
 
 | |
5MI6
 
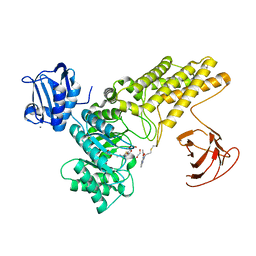 | | BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
8EY1
 
 | |
6UO1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA (containing pseudouridine at the first position of the codon) and deacylated A-, P-, and E-site tRNAs at 2.95A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Dobosz-Bartoszek, M, Polikanov, Y.S. | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Pseudouridinylation of mRNA coding sequences alters translation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8IGF
 
 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with 4-Acetylphenylboronic acid at 2.6 A Resolution | | Descriptor: | (4-ethanoylphenyl)boronic acid, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rasheed, S, Huda, N, Fisher, S.Z, Falke, S, Gul, S, Ahmad, M.S, Choudhary, M.I. | | Deposit date: | 2023-02-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification, crystallization, and first X-ray structure analyses of phenyl boronic acid-based inhibitors of human carbonic anhydrase-II.
Int.J.Biol.Macromol., 267, 2024
|
|
8EQV
 
 | | Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Boudes, M, Zhang, Q, Flanigan, S.F, Davidovich, C. | | Deposit date: | 2022-10-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | To be updated
To Be Published
|
|
5MKM
 
 | |
6Y79
 
 | | Cryo-EM structure of a respiratory complex I F89A mutant | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I.
Nat Commun, 11, 2020
|
|
5MSU
 
 | |
5C0O
 
 | | m1A58 tRNA methyltransferase mutant - Y78A | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Degut, C, Ponchon, L, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
4XT1
 
 | | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Fractalkine, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
6YND
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
5MJJ
 
 | |
