5TN3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S)-17-((4-isopropylphenyl)amino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | Descriptor: | (9beta,13alpha,17beta)-17-{[4-(propan-2-yl)phenyl]amino}estra-1(10),2,4-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6XPT
 
 | |
6XP4
 
 | |
6XPS
 
 | |
3TZE
 
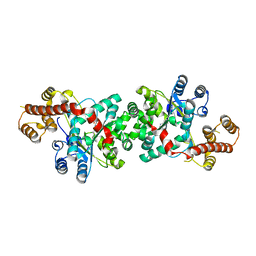 | |
6XPW
 
 | |
6XRQ
 
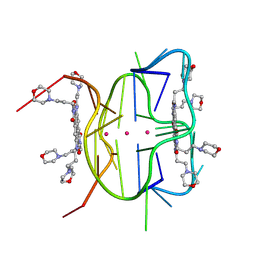 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | Descriptor: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
6CDL
 
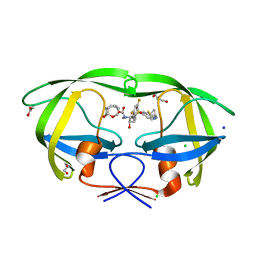 | | HIV-1 wild type protease with GRL-03214A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aR,4S,4aR,7aR,7bR)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
1L5G
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN AVB3 IN COMPLEX WITH AN ARG-GLY-ASP LIGAND | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
7ONI
 
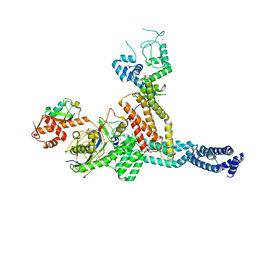 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
7UX4
 
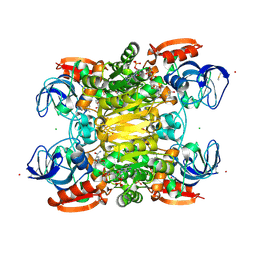 | |
6CDJ
 
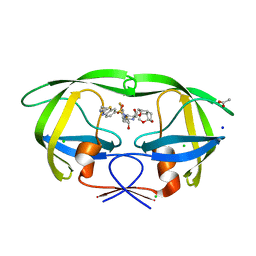 | | HIV-1 wild type protease with GRL-03314A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aS,4R,4aS,7aS,7bS)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
2OH9
 
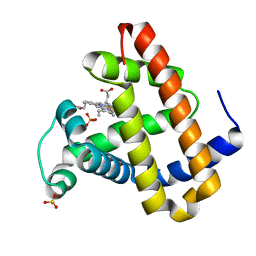 | | Myoglobin cavity mutant V68W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Li, T, Brucker, E.A, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2OHB
 
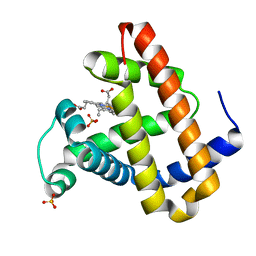 | | Myoglobin cavity mutant I107W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Schweers, R.L, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
2WEY
 
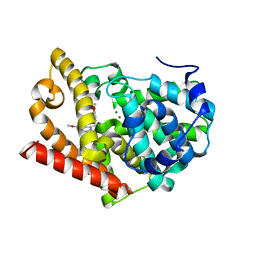 | | Human PDE-papaverine complex obtained by ligand soaking of cross- linked protein crystals | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Andersen, O.A, Schonfeld, D.L, Toogood-Johnson, I, Felicetti, B, Albrecht, C, Fryatt, T, Whittaker, M, Hallett, D, Barker, J. | | Deposit date: | 2009-04-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cross-Linking of Protein Crystals as an Aid in the Generation of Binary Protein-Ligand Crystal Complexes, Exemplified by the Human Pde10A-Papaverine Structure.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1K4W
 
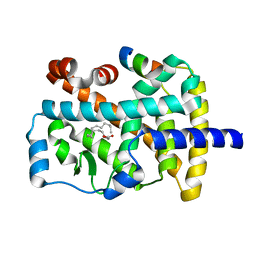 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | Descriptor: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | Authors: | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
5EGV
 
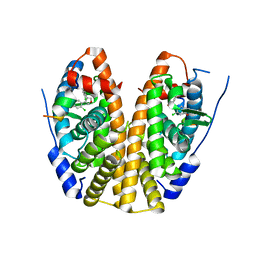 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex the 3,4-diaryl-furan derivative 3-chloranyl-4-[4-(2-chloranyl-4-oxidanyl-phenyl)furan-3-yl]phenol | | Descriptor: | 3-chloranyl-4-[4-(2-chloranyl-4-oxidanyl-phenyl)furan-3-yl]phenol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-10-27 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.863 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5EI1
 
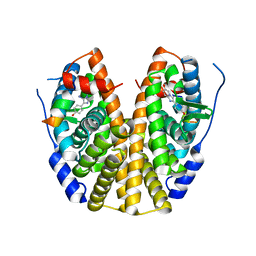 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the imidazopyridine derivative 2-(4-hydroxyphenyl)-3-iodanyl-imidazo[1,2-a]pyridin-6-ol | | Descriptor: | 2-(4-hydroxyphenyl)-3-iodanyl-imidazo[1,2-a]pyridin-6-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-10-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
8CDK
 
 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
6XWE
 
 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
5XGQ
 
 | |
8HUK
 
 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUQ
 
 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
