4EMZ
 
 | |
5OO7
 
 | |
1ZSH
 
 | | Crystal structure of bovine arrestin-2 in complex with inositol hexakisphosphate (IP6) | | Descriptor: | Beta-arrestin 1, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION | | Authors: | Milano, S.K, Kim, Y.M, Stefano, F.P, Benovic, J.L, Brenner, C. | | Deposit date: | 2005-05-24 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding
J.Biol.Chem., 281, 2006
|
|
2PR9
 
 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2QV2
 
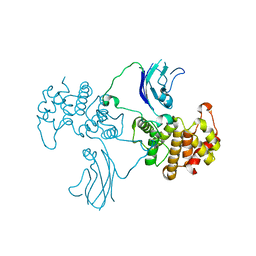 | |
6BNT
 
 | |
3I00
 
 | | Crystal Structure of the huntingtin interacting protein 1 coiled coil domain | | Descriptor: | Huntingtin-interacting protein 1 | | Authors: | Wilbur, J.D, Hwang, P.K, Brodsky, F.M, Fletterick, R.J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accommodation of structural rearrangements in the huntingtin-interacting protein 1 coiled-coil domain.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4UUK
 
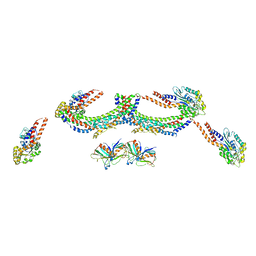 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP strand 2 | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
4UUD
 
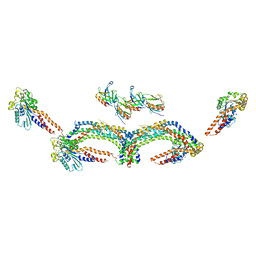 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
1B9K
 
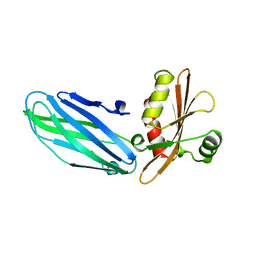 | |
2N12
 
 | |
6A9Y
 
 | | The crystal structure of Mu homology domain of SGIP1 | | Descriptor: | SH3-containing GRB2-like protein 3-interacting protein 1 | | Authors: | Feng, Y, Liu, X. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SGIP1 dimerizes via intermolecular disulfide bond in mu HD domain during cellular endocytosis.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5LP0
 
 | |
5UCC
 
 | | Crystal structure of the ENTH domain of ENT2 from Candida albicans | | Descriptor: | CHLORIDE ION, CITRIC ACID, Potential epsin-like clathrin-binding protein | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the ENTH domain of ENT2 from Candida albicans
To Be Published
|
|
3TJZ
 
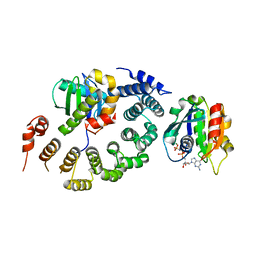 | | Crystal Structure of Arf1 Bound to the gamma/zeta-COP Core Complex | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit gamma, Coatomer subunit zeta-1, ... | | Authors: | Goldberg, J, Yu, X, Breitman, M. | | Deposit date: | 2011-08-25 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structure-based mechanism for arf1-dependent recruitment of coatomer to membranes.
Cell(Cambridge,Mass.), 148, 2012
|
|
3H8D
 
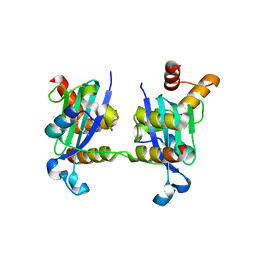 | | Crystal structure of Myosin VI in complex with Dab2 peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Disabled homolog 2, ... | | Authors: | Yu, C, Feng, W, Wei, Z, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
1WR6
 
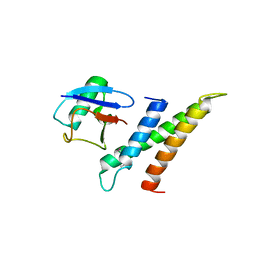 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1NAF
 
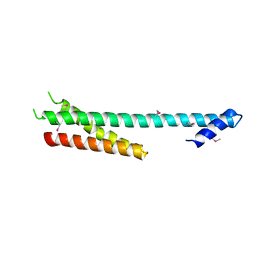 | |
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
1R0D
 
 | |
5AWR
 
 | |
5AWT
 
 | |
5AWU
 
 | |
5AWS
 
 | |
5NZR
 
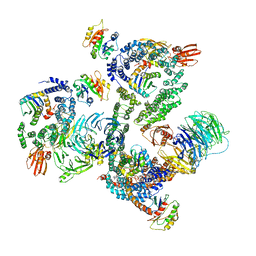 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
