7TGK
 
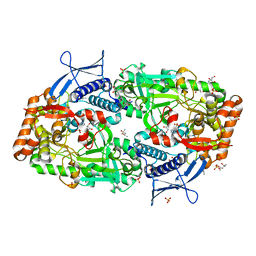 | | Crystal structure of ATP bound DesD, the desferrioxamine synthetase from the Streptomyces griseoflavus ferrimycin biosynthetic pathway | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
7TGN
 
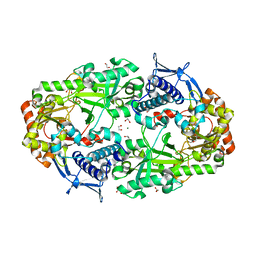 | | Crystal structure of DesD, the desferrioxamine synthetase from the Streptomyces violaceus salmycin biosynthetic pathway | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Desferrioxamine synthetase DesD, ... | | Authors: | Patel, K.D, Gulick, A.M. | | Deposit date: | 2022-01-07 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An acyl-adenylate mimic reveals the structural basis for substrate recognition by the iterative siderophore synthetase DesD.
J.Biol.Chem., 298, 2022
|
|
3T3I
 
 | | Glycogen Phosphorylase b in complex with GlcCF3U | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
7TSW
 
 | |
3RPL
 
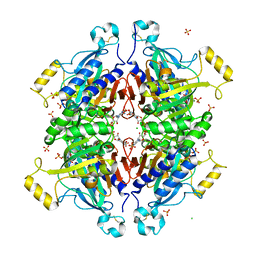 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
7TNV
 
 | |
7THG
 
 | |
3T3E
 
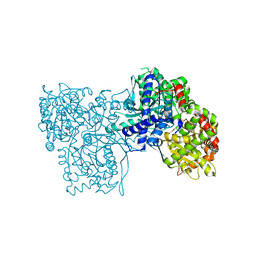 | | Glycogen phosphorylase b in complex with GlcClU | | Descriptor: | 5-chloro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
7TGM
 
 | |
7TGJ
 
 | |
8QSX
 
 | |
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
3T3H
 
 | | Glycogen Phosphorylase b in complex with GlcIU | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-iodopyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
6U3E
 
 | |
3T3G
 
 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3D
 
 | | Glycogen phosphorylase b in complex with GlcU | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3S0J
 
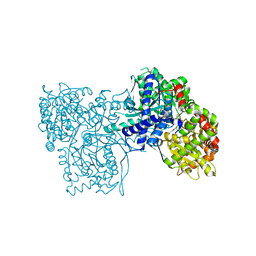 | | The crystal structure of glycogen phosphorylase b in complex with 2,5-dihydroxy-4-(beta-D-glucopyranosyl)-chlorobenzene | | Descriptor: | (1S)-1,5-anhydro-1-(4-chloro-2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, muscle form | | Authors: | Alexacou, K.-M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G, Leonidas, D.D. | | Deposit date: | 2011-05-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogen-substituted (C-beta-D-glucopyranosyl)-hydroquinone regioisomers: synthesis, enzymatic evaluation and their binding to glycogen phosphorylase.
Bioorg.Med.Chem., 19, 2011
|
|
3PRV
 
 | | Nucleoside diphosphate kinase B from Trypanosoma cruzi | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Souza, T.A.C.B, Trindade, D.M, Tonoli, C.C.C, Santos, C.R, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-11-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3TZE
 
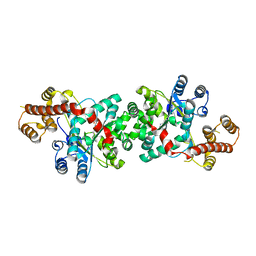 | |
3QTY
 
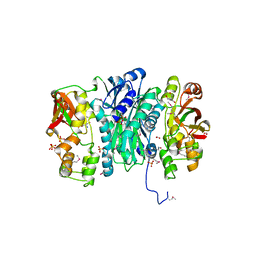 | | Crystal structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis complexed with pyrophosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis complexed with pyrophosphate
To be Published
|
|
3U39
 
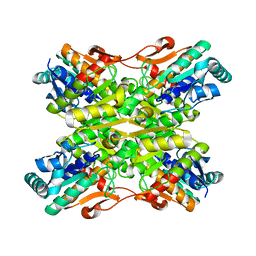 | | Crystal Structure of the apo Bacillus Stearothermophilus phosphofructokinase | | Descriptor: | 6-phosphofructokinase, CALCIUM ION | | Authors: | Mosser, R, Reddy, M.C.M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7921 Å) | | Cite: | Structure of the apo form of Bacillus stearothermophilus phosphofructokinase.
Biochemistry, 51, 2012
|
|
3U53
 
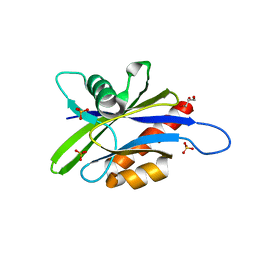 | | Crystal structure of human Ap4A hydrolase | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, SULFATE ION | | Authors: | Ge, H.H, Teng, M.K. | | Deposit date: | 2011-10-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4G0P
 
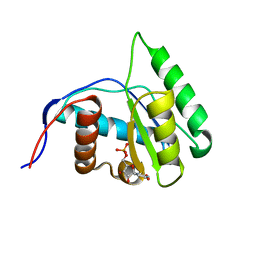 | | Crystal Structure of Arabidopsis thaliana AGO1 MID domain in complex with UMP | | Descriptor: | Protein argonaute 1, URIDINE-5'-MONOPHOSPHATE | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
2QN8
 
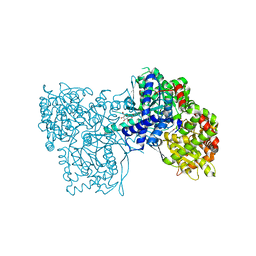 | | Glycogen Phosphorylase b in complex with N-4-nitrobenzoyl-N'-beta-D-glucopyranosyl urea | | Descriptor: | Glycogen phosphorylase, muscle form, N-{[(4-nitrophenyl)carbonyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Tiraidis, C, Alexacou, K.-M, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-(4-substituted-benzoyl)-N'-(beta-D-glucopyranosyl)ureas, inhibitors of glycogen phosphorylase: synthesis, kinetic and crystallographic evaluation
To be Published
|
|
2R7Z
 
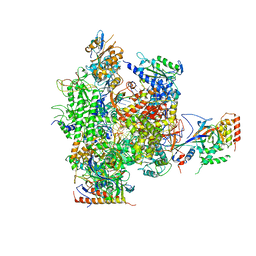 | | Cisplatin lesion containing RNA polymerase II elongation complex | | Descriptor: | 5'-D(*CP*AP*AP*GP*TP*AP*G)-3', 5'-D(*TP*AP*CP*TP*TP*GUP*CP*CP*CP*TP*CP*CP*TP*CP*AP*T)-3', 5'-R(*UP*UP*UP*GP*AP*GP*GP*AP*GP*G)-3', ... | | Authors: | Damsma, G.E, Alt, A, Brueckner, F, Carell, T, Cramer, P. | | Deposit date: | 2007-09-10 | | Release date: | 2007-11-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of transcriptional stalling at cisplatin-damaged DNA.
Nat.Struct.Mol.Biol., 14, 2007
|
|
