6VET
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5E0G
 
 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (17-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(8~{S},11~{S},14~{S})-8-(hydroxymethyl)-11-(2-methylpropyl)-5,10,13-tris(oxidanylidene)-1,4,9,12,17,18-hexazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
6PEA
 
 | | High Resolution Apo Carbonic Anhydrase II | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2 | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2019-06-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and mechanism of copper-carbonic anhydrase II: a nitrite reductase.
Iucrj, 7, 2020
|
|
6GB6
 
 | | Structure of H-2Kb with dipeptide GL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5VQ4
 
 | | Nitrogenase Av1 at pH 5 | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Spatzal, T, Rees, D.C. | | Deposit date: | 2017-05-07 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reversible Protonated Resting State of the Nitrogenase Active Site.
J. Am. Chem. Soc., 139, 2017
|
|
6PEX
 
 | |
6GBQ
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Reston virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6YII
 
 | |
5E1M
 
 | | Crystal structure of NTMT1 in complex with PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
6FWA
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 7-Methyl-L-Tryptophan | | Descriptor: | 7-Methyl-L-Tryptophan, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.846 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
5KOF
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, [(3~{S})-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-(prop-2-enoylamino)ethylamino]propyl]amino]butyl] dihydrogen phosphate | | Authors: | Milligan, J.C, Jackson, D.R, Barajas, J.F, Tsai, S.C. | | Deposit date: | 2016-06-30 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for interactions between an acyl carrier protein and a ketosynthase.
Nat.Chem.Biol., 15, 2019
|
|
5E4P
 
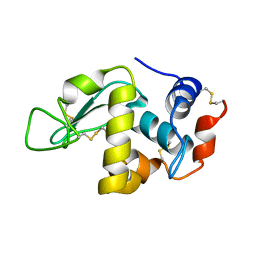 | | X-ray Crystal Structure Analysis of Magnetically Oriented Microcrystals of Lysozyme at 1.8 angstrom Resolution | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Garman, E.F, Baba, S, Mizuno, N, Mikami, B, Kimura, T. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | X-ray crystal structure analysis of magnetically oriented microcrystals of lysozyme at 1.8 A resolution
J.Appl.Crystallogr., 49, 2016
|
|
6FWL
 
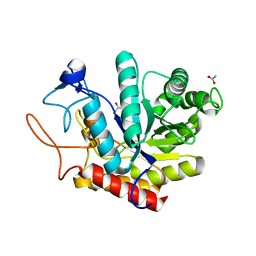 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex alpha-Glc-1,3-(1,2-anhydro-carba-mannose) | | Descriptor: | (1~{S},2~{S},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
5KOM
 
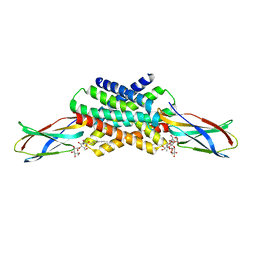 | | The crystal structure of fluoride channel Fluc Ec2 F83I Mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, Putative fluoride ion transporter CrcB, ... | | Authors: | Last, N.B, Kolmakova-Partensky, L, Shane, T, Miller, C. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanistic signs of double-barreled structure in a fluoride ion channel.
Elife, 5, 2016
|
|
5HY2
 
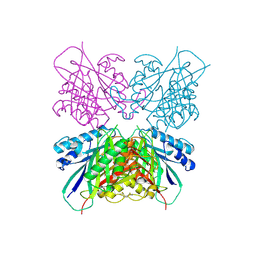 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
7CLU
 
 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
6FWP
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose | | Descriptor: | ACETATE ION, Glycosyl hydrolase family 71, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6N8S
 
 | |
8OTN
 
 | | structure of InhA from mycobacterium tuberculosis in complex with inhibitor 7-((1-(3-Hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-4-methyl-2H-chromen-2-one | | Descriptor: | 4-methyl-7-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methoxy]chromen-2-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
6UXL
 
 | |
6PGA
 
 | |
5E2A
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
6G48
 
 | | Sporosarcina pasteurii urease inhibited by silver | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-03-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of urease inactivated by Ag(i): a new paradigm for enzyme inhibition by heavy metals.
Dalton Trans, 47, 2018
|
|
6N9B
 
 | | FtsY-NG ultra high-resolution | | Descriptor: | ACETATE ION, Signal recognition particle receptor FtsY | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
6G4F
 
 | |
