1ABJ
 
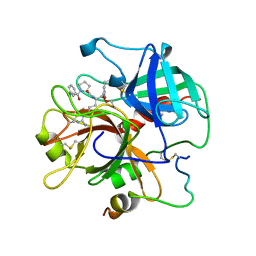 | |
1FOD
 
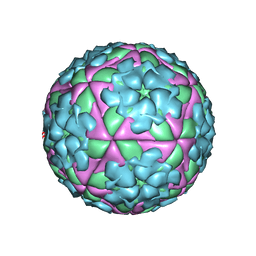 | | STRUCTURE OF A MAJOR IMMUNOGENIC SITE ON FOOT-AND-MOUTH DISEASE VIRUS | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS | | Authors: | Logan, D.T, Lea, S, Lewis, R, Stuart, D, Fry, E. | | Deposit date: | 1993-10-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a major immunogenic site on foot-and-mouth disease virus.
Nature, 362, 1993
|
|
1BSC
 
 | |
1CCD
 
 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
1FKS
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1SMR
 
 | |
1SCB
 
 | | ENZYME CRYSTAL STRUCTURE IN A NEAT ORGANIC SOLVENT | | Descriptor: | ACETONITRILE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Fitzpatrick, P.A, Steinmetz, A.C.U, Ringe, D, Klibanov, A.M. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme crystal structure in a neat organic solvent.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1AAF
 
 | | NUCLEOCAPSID ZINC FINGERS DETECTED IN RETROVIRUSES: EXAFS STUDIES ON INTACT VIRUSES AND THE SOLUTION-STATE STRUCTURE OF THE NUCLEOCAPSID PROTEIN FROM HIV-1 | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Henderson, L.E, Chance, M.R, Bess Junior, J.W, South, T.L, Blake, P.R, Sagi, I, Perez-Alvarado, G, Sowder, R.C, Hare, D.R, Arthur, L.O. | | Deposit date: | 1992-04-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1.
Protein Sci., 1, 1992
|
|
1ADG
 
 | | CRYSTALLOGRAPHIC STUDIES OF TWO ALCOHOL DEHYDROGENASE-BOUND ANALOGS OF THIAZOLE-4-CARBOXAMIDE ADENINE DINUCLEOTIDE (TAD), THE ACTIVE ANABOLITE OF THE ANTITUMOR AGENT TIAZOFURIN | | Descriptor: | ALCOHOL DEHYDROGENASE, BETA-METHYLENE-SELENAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, ZINC ION | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Marquez, V.E, Carrell, H.L, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry, 33, 1994
|
|
1ABN
 
 | |
1TAS
 
 | |
2BAT
 
 | | THE STRUCTURE OF THE COMPLEX BETWEEN INFLUENZA VIRUS NEURAMINIDASE AND SIALIC ACID, THE VIRAL RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor.
Proteins, 14, 1992
|
|
1KAA
 
 | |
2BBM
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
1SUD
 
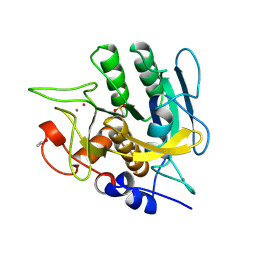 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
2BBN
 
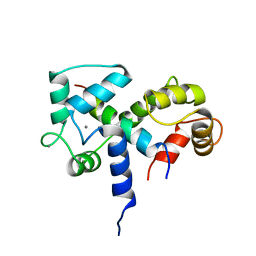 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2AT2
 
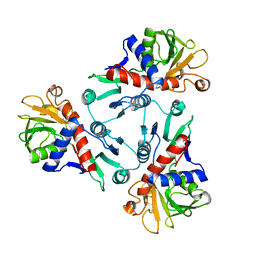 | |
1TRB
 
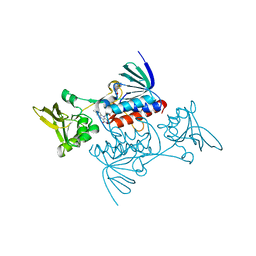 | |
2BBQ
 
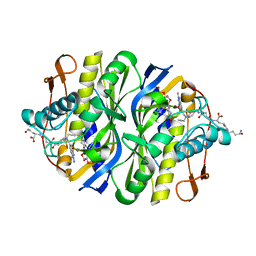 | | STRUCTURAL BASIS FOR RECOGNITION OF POLYGLUTAMYL FOLATES BY THYMIDYLATE SYNTHASE | | Descriptor: | 10-PARPARGYL-5,8-DIDEAZAFOLATE-4-GLUTAMIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Kamb, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1992-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of polyglutamyl folates by thymidylate synthase.
Biochemistry, 31, 1992
|
|
2BBK
 
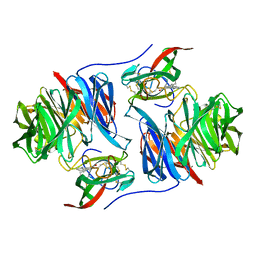 | |
1CAO
 
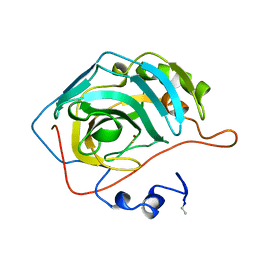 | |
1CBV
 
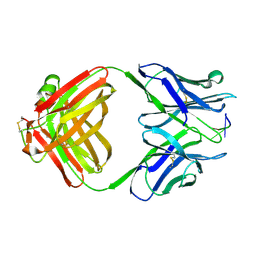 | | AN AUTOANTIBODY TO SINGLE-STRANDED DNA: COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF THE UNLIGANDED FAB AND A DEOXYNUCLEOTIDE-FAB COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (FAB (BV04-01) AUTOANTIBODY-HEAVY CHAIN), PROTEIN (FAB (BV04-01) AUTOANTIBODY-LIGHT CHAIN) | | Authors: | Herron, J.N, He, X.M, Ballard, D.W, Blier, P.R, Pace, P.E, Bothwell, A.L.M, Voss Junior, E.W, Edmundson, A.B. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | An autoantibody to single-stranded DNA: comparison of the three-dimensional structures of the unliganded Fab and a deoxynucleotide-Fab complex.
Proteins, 11, 1991
|
|
1TYP
 
 | |
1FPH
 
 | |
1CDG
 
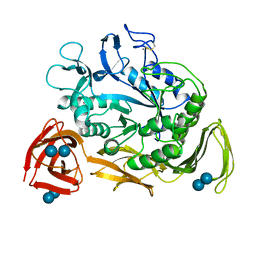 | | NUCLEOTIDE SEQUENCE AND X-RAY STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 IN A MALTOSE-DEPENDENT CRYSTAL FORM | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLYCOSYL-TRANSFERASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lawson, C.L, Van Montfort, R, Strokopytov, B.V, Kalk, K.H, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1993-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form.
J.Mol.Biol., 236, 1994
|
|
