2ZCJ
 
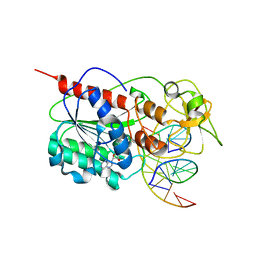 | | Ternary structure of the Glu119Gln M.HhaI, C5-Cytosine DNA methyltransferase, with unmodified DNA and AdoHcy | | Descriptor: | DNA (5'-D(*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), Modification methylase HhaI, ... | | Authors: | Shieh, F.K, Reich, N.O. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | AdoMet-dependent Methyl-transfer: Glu(119) Is Essential for DNA C5-Cytosine Methyltransferase M.HhaI
J.Mol.Biol., 373, 2007
|
|
6LQP
 
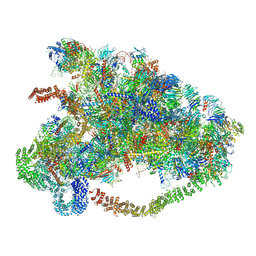 | |
4V6U
 
 | | Promiscuous behavior of proteins in archaeal ribosomes revealed by cryo-EM: implications for evolution of eukaryotic ribosomes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10P, ... | | Authors: | Armache, J.-P, Anger, A.M, Marquez, V, Frankenberg, S, Froehlich, T, Villa, E, Berninghausen, O, Thomm, M, Arnold, G.J, Beckmann, R, Wilson, D.N. | | Deposit date: | 2012-08-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution.
Nucleic Acids Res., 41, 2013
|
|
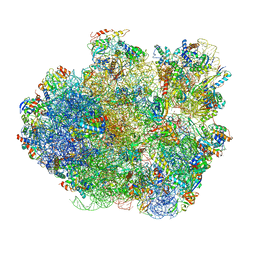
4V6X
 
 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
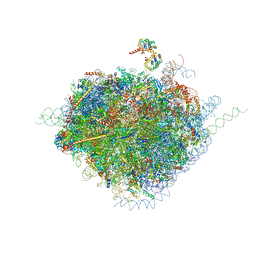
9B8M
 
 | | Crystal structure of ornithine decarboxylase in complex with a novel inhibitor | | Descriptor: | O-{[(3R)-pyrrolidin-3-yl]methyl}hydroxylamine, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schultz, C.R, Aleiwi, B, Zhou, X.E, Suino-Powell, K, Melcher, K, Brunzelle, J.S, Almeida, N.M.S, Wilson, A.K, Ellsworth, E, Bachmann, A.S. | | Deposit date: | 2024-03-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Activity of Novel Ornithine Decarboxylase (ODC) Inhibitors.
J.Med.Chem., 68, 2025
|
|
9B8N
 
 | | Crystal structure of ornithine decarboxylase in complex with a novel inhibitor (10-S) | | Descriptor: | O-{[(3S)-pyrrolidin-3-yl]methyl}hydroxylamine, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schultz, C.R, Aleiwi, B, Zhou, X.E, Suino-Powell, K, Melcher, K, Brunzelle, J.S, Almeida, N.M.S, Wilson, A.K, Ellsworth, E, Bachmann, A.S. | | Deposit date: | 2024-03-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Biological Activity of Novel Ornithine Decarboxylase (ODC) Inhibitors.
J.Med.Chem., 68, 2025
|
|
3ZCX
 
 | |
6LQU
 
 | |
6P5N
 
 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
3X2S
 
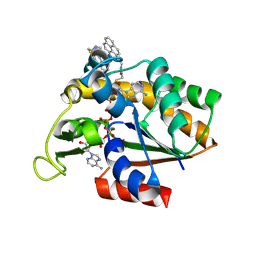 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4V4N
 
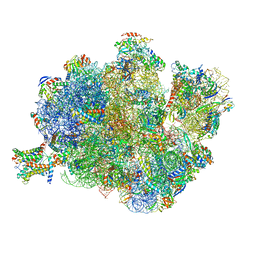 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
9BUP
 
 | |
4QBG
 
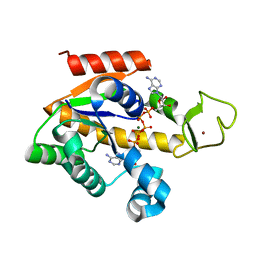 | | Crystal structure of a stable adenylate kinase variant AKlse4 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
1QBB
 
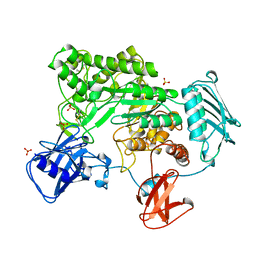 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBA
 
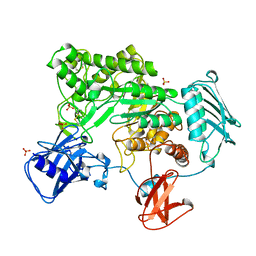 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QF9
 
 | |
6P5K
 
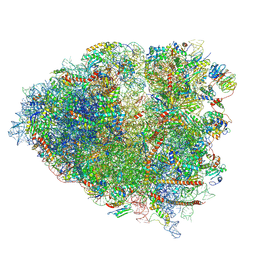 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
4QBH
 
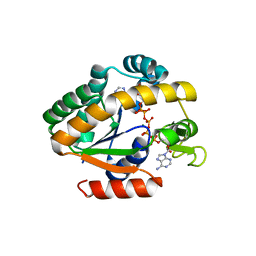 | | Crystal structure of a stable adenylate kinase variant AKlse5 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2014-05-08 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
7QEP
 
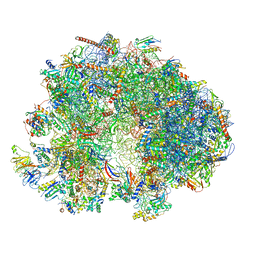 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
9C8K
 
 | |
3A28
 
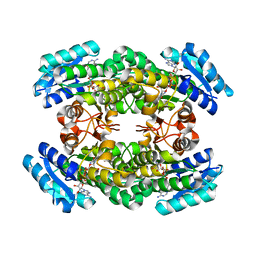 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
6MHT
 
 | |
5NDG
 
 | | Crystal structure of geneticin (G418) bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6UZ7
 
 | | K.lactis 80S ribosome with p/PE tRNA and eIF5B | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Fernandez, I.S, Huang, B.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
