3G1R
 
 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | Descriptor: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
3GJ8
 
 | |
6NE5
 
 | |
8CIM
 
 | | BA.2-07 FAB IN COMPLEX WITH SARS-COV-2 BA.2.12.1 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.2-07 FAB HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Isolation of a pair of potent broadly neutralizing mAb binding to RBD and SD1 domains of SARS-CoV-2
Res Sq, 2023
|
|
6AUC
 
 | | Artificial metalloproteins containing a Co4O4 active site - 2xm-Sav | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)4(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurackal, J, Huerta-Lavorie, R, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6AN8
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6SSO
 
 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
6NFV
 
 | | Structure of the KcsA-G77C mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AUS
 
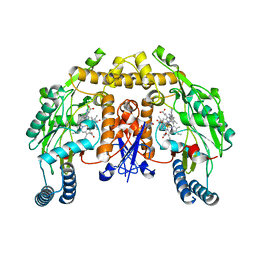 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(3-(methylamino)propyl)benzonitrile | | Descriptor: | 3-[2-(6-amino-4-methylpyridin-2-yl)ethyl]-5-[3-(methylamino)propyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6AV6
 
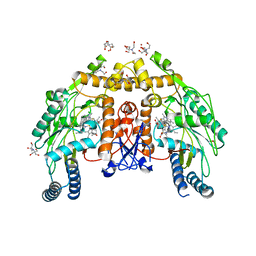 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-Fluoro-5-(3-(methylamino)propyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-fluoro-5-[3-(methylamino)propyl]phenyl}ethyl)-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6M66
 
 | | The Cryo-EM Structure of Human Pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M79
 
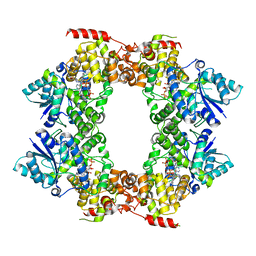 | |
4DNE
 
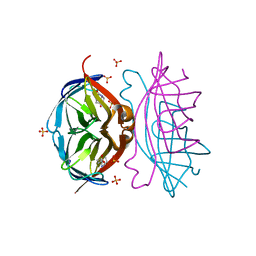 | | Crystal structure of a triple-mutant of streptavidin in complex with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, SULFATE ION, Streptavidin | | Authors: | Panwar, P, Deniaud, A, Pebay-Peyroula, E. | | Deposit date: | 2012-02-08 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Contamination from an affinity column: an encounter with a new villain in the world of membrane-protein crystallization.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3G37
 
 | | Cryo-EM structure of actin filament in the presence of phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wakabayshi, T, Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q. | | Deposit date: | 2009-02-02 | | Release date: | 2010-11-03 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3I2K
 
 | | Cocaine Esterase, wild type, bound to a DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
3I3E
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | Deposit date: | 2009-06-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
6TCL
 
 | | Photosystem I tetramer | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
3G5F
 
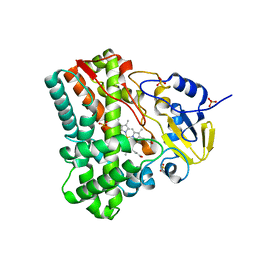 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5U
 
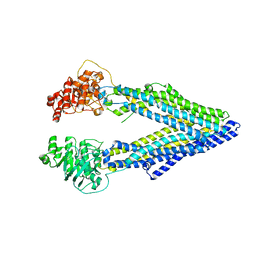 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
6DDU
 
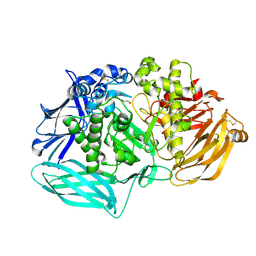 | | mouse beta-mannosidase bound to beta-D-mannose (MANBA) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2018-05-10 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.668 Å) | | Cite: | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
3G73
 
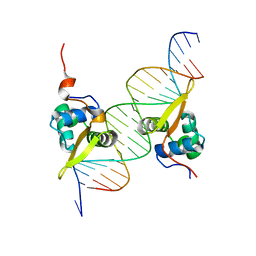 | | Structure of the FOXM1 DNA binding | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(P*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein M1, ... | | Authors: | Littler, D.R, Perrakis, A, Hibbert, R.G, Medema, R.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence
Nucleic Acids Res., 2010
|
|
3I53
 
 | |
3I4D
 
 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
