3VOR
 
 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VN5
 
 | | Crystal structure of Aquifex aeolicus RNase H3 | | Descriptor: | Ribonuclease HIII | | Authors: | Jongruja, N, You, D.J, Eiko, K, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and characterization of RNase H3 from Aquifex aeolicus
Febs J., 279, 2012
|
|
3UTQ
 
 | | Human HLA-A*0201-ALWGPDPAAA | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
3UC7
 
 | | Trp-cage cyclo-TC1 - monoclinic crystal form | | Descriptor: | CHLORIDE ION, Cyclo-TC1 | | Authors: | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UI5
 
 | | Crystal structure of human Parvulin 14 | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
3UC9
 
 | |
3UKA
 
 | |
3ULT
 
 | | Crystal structure of an ice-binding protein from the perennial ryegrass, Lolium perenne | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Ice recrystallization inhibition protein-like protein | | Authors: | Middleton, A.J, Faucher, F, Campbell, R.L, Davies, P.L. | | Deposit date: | 2011-11-11 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antifreeze protein from freeze-tolerant grass has a beta-roll fold with an irregularly structured ice-binding site.
J.Mol.Biol., 416, 2012
|
|
3UDH
 
 | | Crystal Structure of BACE with Compound 1 | | Descriptor: | (3S)-spiro[indole-3,3'-pyrrolidin]-2(1H)-one, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UDQ
 
 | | Crystal Structure of BACE with Compound 13 | | Descriptor: | (4S)-6-bromo-1,1-dioxido-3,4-dihydro-2H-thiochromen-4-yl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, Beta-secretase 1 | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UE2
 
 | |
3UGJ
 
 | | Formyl Glycinamide ribonucletide amidotransferase from Salmonella Typhimurum: Role of the ATP complexation and glutaminase domain in catalytic coupling | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Morar, M, Tanwar, A.S, Panjikar, S, Anand, R. | | Deposit date: | 2011-11-02 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UI6
 
 | | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SODIUM ION, ... | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | 0.89 A resolution crystal structure of human Parvulin 14 in complex with oxidized DTT
To be Published
|
|
3UIN
 
 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UDS
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, MAGNESIUM ION | | Authors: | Gosein, V, Leung, T.-F, Krajden, O, Miller, G.J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inositol phosphate-induced stabilization of inositol 1,3,4,5,6-pentakisphosphate 2-kinase and its role in substrate specificity.
Protein Sci., 21, 2012
|
|
3UJ4
 
 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
3UKP
 
 | |
3UGF
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UMW
 
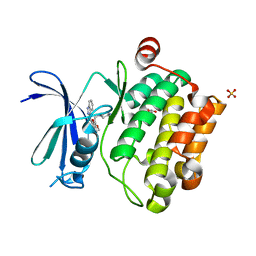 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indazol-3-yl)methylene]-6-methoxy-7-(piperazin-1-ylmethyl)benzofuran-3(2H)-one | | Descriptor: | (2Z)-2-(1H-indazol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3(2H)-one, GLYCEROL, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational evolution of a novel type of potent and selective proviral integration site in Moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound.
J.Med.Chem., 55, 2012
|
|
3UIX
 
 | |
3UR9
 
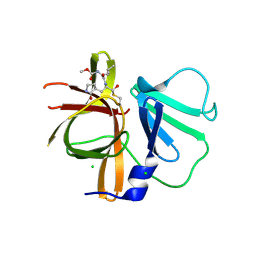 | | 1.65A resolution structure of Norwalk Virus Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like protease, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3UJE
 
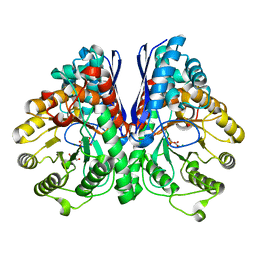 | | Asymmetric complex of human neuron specific enolase-3-PGA/PEP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
3UKH
 
 | |
3UUF
 
 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
3UWT
 
 | |
