5UBU
 
 | | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica. | | Descriptor: | Putative acetamidase/formamidase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.
To Be Published
|
|
4XMT
 
 | |
4XMW
 
 | |
6X1Q
 
 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
6GJF
 
 | | Ancestral endocellulase Cel5A | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ENDOGLUCANASE, ... | | Authors: | Gavira, J.A, Perez-Jimenez, R, Barruetabena-Garate, N. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Resurrection of efficient Precambrian endoglucanases for lignocellulosic biomass hydrolysis
Commun Chem, 2019
|
|
5NI3
 
 | | sfGFP 204-204 mutant dimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Worthy, H.L, Rizkallah, P.J. | | Deposit date: | 2017-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Association of Fluorescent Protein Pairs and Its Significant Impact on Fluorescence and Energy Transfer
Adv Sci, 2020
|
|
6CQP
 
 | | High resolution crystal structure of FtsY-NG domain of E. coli | | Descriptor: | GLYCEROL, SODIUM ION, Signal recognition particle receptor FtsY | | Authors: | Faoro, C, Ataide, S.F. | | Deposit date: | 2018-03-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
4N3P
 
 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
5UQA
 
 | |
4RYF
 
 | | ClpP1/2 heterocomplex from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, MALONATE ION, SODIUM ION | | Authors: | Dahmen, M, Vielberg, M.-T, Groll, M, Sieber, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AGV
 
 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, CYCLOHEXYL GRISELIMYCIN, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
4EQI
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1, SODIUM ION | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
5T73
 
 | |
5DYN
 
 | | B. fragilis cysteine protease | | Descriptor: | CHLORIDE ION, Putative peptidase, SODIUM ION | | Authors: | Choi, V.M, Herrou, J, Hecht, A.L, Turner, J.R, Crosson, S, Bubeck Wardenburg, J. | | Deposit date: | 2015-09-24 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice.
Nat. Med., 22, 2016
|
|
6HJB
 
 | | Xray structure of GLIC in complex with malonate | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for the binding of monocarboxylates and dicarboxylates at pharmacologically relevant extracellular sites of a pentameric ligand-gated ion channel.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4GCV
 
 | |
6LON
 
 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6AM7
 
 | |
5EYU
 
 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
6H6Y
 
 | | GI.1 human norovirus protruding domain in complex with Nano-7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Kilic, T, Ruoff, K, Hansman, G.S. | | Deposit date: | 2018-07-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Nanobodies Targeting the Prototype Norovirus.
J. Virol., 93, 2019
|
|
6M2R
 
 | | X-ray structure of a functional Drosophila dopamine transporter in L-norepinephrine bound form | | Descriptor: | Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-28 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6P8L
 
 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX (Zn Absorption Edge X-ray Data) | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
7D3U
 
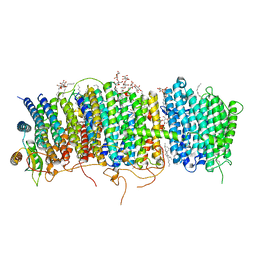 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | Descriptor: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | Authors: | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P8K
 
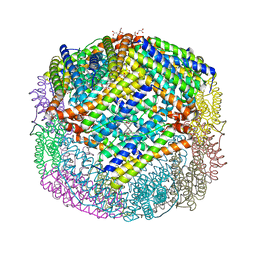 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
6HJ3
 
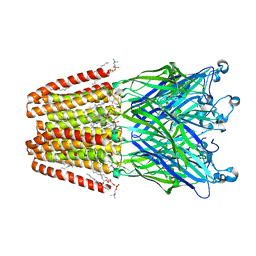 | | Xray structure of GLIC in complex with fumarate | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2018-08-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the binding of monocarboxylates and dicarboxylates at pharmacologically relevant extracellular sites of a pentameric ligand-gated ion channel.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
