8WL9
 
 | |
8WL7
 
 | |
8WBV
 
 | |
6T1D
 
 | | Pleurotus Ostreatus Lectin (POL), compelx with melibiose | | Descriptor: | CALCIUM ION, Lectin, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Destefanis, L, Perduca, M, Bovi, M, Monaco, H.L, Capaldi, S. | | Deposit date: | 2019-10-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and properties of the oyster mushroom (Pleurotus ostreatus) lectin.
Glycobiology, 30, 2020
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6SGV
 
 | | Crystal structure of AcAChBP in complex with hosieine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Hosieine, ... | | Authors: | Hunter, W.N, Dawson, A, Parker, H. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating the activity of the potent nicotinic acetylcholine receptor agonists (+)-anatoxin-a and (-)-hosieine-A
Acta Crystallogr.,Sect.F, 2022
|
|
4MQF
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist 2-hydroxysaclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxysaclofen, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MQE
 
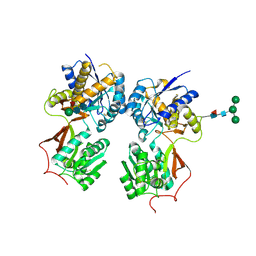 | | Crystal structure of the extracellular domain of human GABA(B) receptor in the apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MR8
 
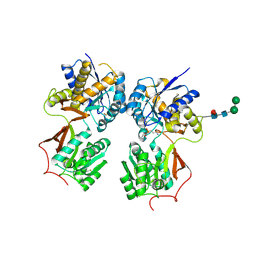 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP35348 | | Descriptor: | (S)-(3-aminopropyl)(diethoxymethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MR7
 
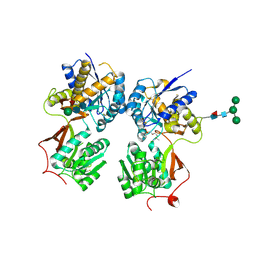 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4N61
 
 | |
6STT
 
 | | Adenovirus 29 Fiber Knob protein in complex with Sialic acid | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
4MS1
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP46381 | | Descriptor: | (S)-(3-aminopropyl)(cyclohexylmethyl)phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4N62
 
 | |
4N60
 
 | |
4MR9
 
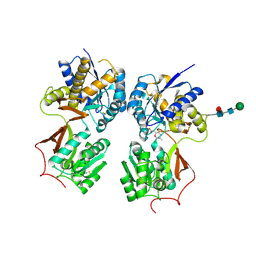 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist SCH50911 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4N63
 
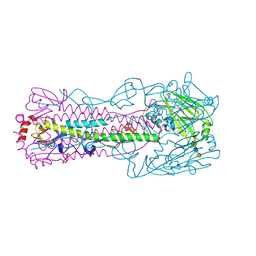 | |
4N5K
 
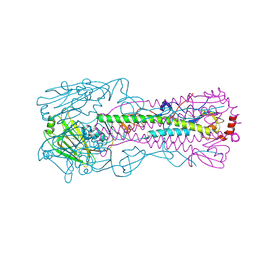 | |
8VFT
 
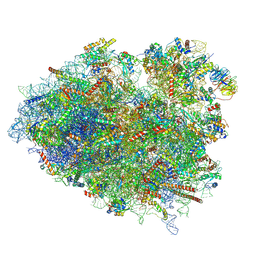 | |
6ZOG
 
 | | Minocycline binding to the deep binding pocket of AcrB-I38F_I671T | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 1,2-ETHANEDIOL, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
6ZOE
 
 | | AcrB-F563A symmetric T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
6ZO6
 
 | | Minocycline binding to the deep binding pocket of AcrB-G619P | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
8VGY
 
 | | Crystal structure of human apoptosis-inducing factor (AIF) bound to the fused N-terminal domain of CHCHD4 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis-inducing factor 1, mitochondrial,Mitochondrial intermembrane space import and assembly protein 40, ... | | Authors: | Brosey, C.A, Tainer, J.A. | | Deposit date: | 2023-12-29 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
Embo J., 44, 2025
|
|
6ZOA
 
 | | Partially induced AcrB T protomer and DDM binding to the TM8/PC2 pathway of AcrB L2 protomer | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPIN, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
