6L22
 
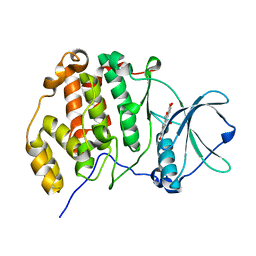 | | Crystal structure of CK2a1 H115Y with hematein | | Descriptor: | (6aR)-3,4,6a,10-tetrakis(oxidanyl)-6,7-dihydroindeno[2,1-c]chromen-9-one, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12318087 Å) | | Cite: | Structural insights for producing CK2 alpha 1-specific inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LJ0
 
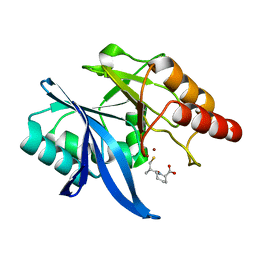 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss02122 | | Descriptor: | (2R)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-2-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJB
 
 | |
2IKF
 
 | |
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2W
 
 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | Descriptor: | freshwater cyanophage protein | | Authors: | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-10-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
2IMB
 
 | |
6L34
 
 | |
2IMT
 
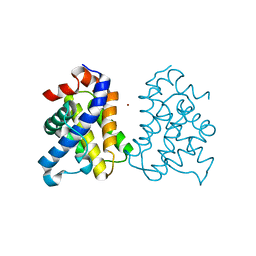 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site.
Mol.Cell, 24, 2006
|
|
6L3F
 
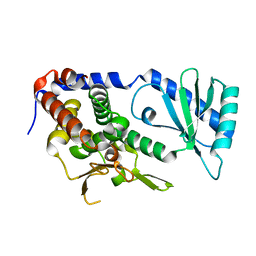 | |
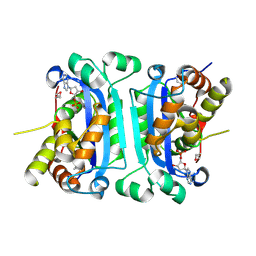
2IOC
 
 | |
6LJQ
 
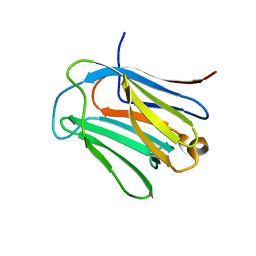 | | human galectin-16 R55N | | Descriptor: | Galectin-16, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6L4R
 
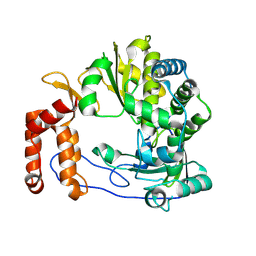 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
6L50
 
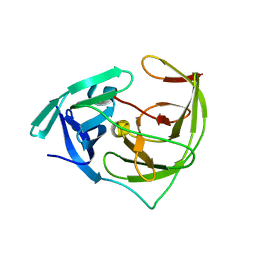 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 2-sulfanylidene-1,3-thiazolidin-4-one, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
2ICH
 
 | |
6L5S
 
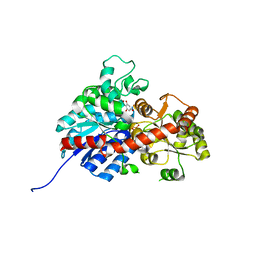 | | crystal structure of GgCGT in complex with UDP-Glu | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, GLYCEROL, GgCGT, ... | | Authors: | Zhang, M, Li, F.D, Ye, M. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Functional Characterization and Structural Basis of an Efficient Di-C-glycosyltransferase fromGlycyrrhiza glabra.
J.Am.Chem.Soc., 142, 2020
|
|
2ID4
 
 | |
6LK5
 
 | | MLKL mutant - T357ES358D | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H.Y, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
2IPM
 
 | |
6KZ1
 
 | | Complex structure of Whirlin and Myosin XVa | | Descriptor: | Myosin XVa, Whirlin | | Authors: | Lin, L, Wang, M, Shi, Y, Zhu, J, Zhang, R. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex.
Cell Rep, 34, 2021
|
|
6KZ6
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
2IQJ
 
 | | Crystal structure of the GAP domain of SMAP1L (LOC64744) stromal membrane-associated protein 1-like | | Descriptor: | BETA-MERCAPTOETHANOL, Stromal membrane-associated protein 1-like, UNKNOWN ATOM OR ION, ... | | Authors: | Tong, Y, Dimov, S, Shen, L, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-13 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the GAP domain of SMAP1L (LOC64744) stromal membrane-associated protein 1-like
To be Published
|
|
6L6C
 
 | | X-ray structure of human galectin-10 in complex with D-arabinose | | Descriptor: | Galectin-10, alpha-D-arabinopyranose | | Authors: | Kamitori, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
