6L6Q
 
 | |
6LAX
 
 | | the mutant SAM-VI riboswitch (U6C) bound to SAM | | Descriptor: | RNA (55-MER), S-ADENOSYLMETHIONINE, U1 small nuclear ribonucleoprotein A | | Authors: | Sun, A, Ren, A. | | Deposit date: | 2019-11-13 | | Release date: | 2020-01-01 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAM-VI riboswitch structure and signature for ligand discrimination.
Nat Commun, 10, 2019
|
|
6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
6LC7
 
 | | Crystal structure of AmpC Ent385 free form | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Beta-lactamase, GLYCEROL, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
6LD5
 
 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, RNA-directed RNA polymerase NS5, ZINC ION, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6LDS
 
 | |
6LE4
 
 | |
6LF1
 
 | | SeviL, a GM1b/asialo-GM1 binding lectin | | Descriptor: | CHLORIDE ION, SeviL | | Authors: | Kamata, K, Ozeki, Y, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2019-11-28 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of SeviL, a GM1b/asialo-GM1 binding R-type lectin from the mussel Mytilisepta virgata.
Sci Rep, 10, 2020
|
|
6L74
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 2 nt | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-10-31 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L8B
 
 | | The ligand-free structure of human PPARgamma LBD | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, D.M, Han, B.W. | | Deposit date: | 2019-11-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Cyclin-Dependent Kinase 5 Inhibitor Butyrolactone I Elicits a Partial Agonist Activity of Peroxisome Proliferator-Activated Receptor gamma.
Biomolecules, 10, 2020
|
|
6LF3
 
 | |
2INZ
 
 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
6LGM
 
 | |
6LHJ
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with C452 (compound 16) and NADPH | | Descriptor: | 1-[3-(2-chloranyl-4-fluoranyl-phenoxy)propoxy]-6,6-dimethyl-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-12-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
2IO5
 
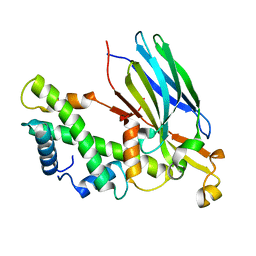 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
6L8J
 
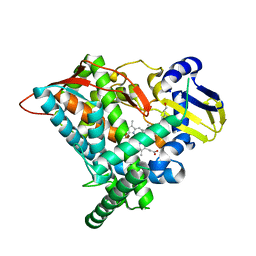 | | Crystal structure of CYP97A3 mutant S290D/W300L/S304V in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Liu, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2IOD
 
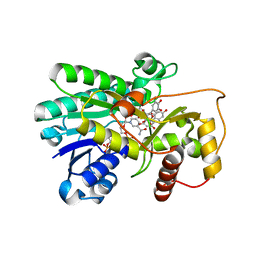 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois d'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
2IPI
 
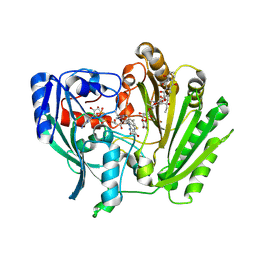 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H -PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-C ARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6L8Q
 
 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
6LB9
 
 | | Magnesium ion-bound SspB crystal structure | | Descriptor: | DUF4007 domain-containing protein, MAGNESIUM ION | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
6LBJ
 
 | |
6LC1
 
 | |
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LIG
 
 | |
2IT9
 
 | |
