6XJF
 
 | |
8PBL
 
 | |
4ZRL
 
 | | Structure of the non canonical Poly(A) polymerase complex GLD-2 - GLD-3 | | Descriptor: | CHLORIDE ION, Defective in germ line development protein 3, Poly(A) RNA polymerase gld-2 | | Authors: | Nakel, K, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2015-05-12 | | Release date: | 2015-07-08 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6H80
 
 | | Dengue-RdRp3-inhibitor complex co-crystallisation | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6H9R
 
 | | Dengue-RdRp3-inhibitor complex soaking | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6SZU
 
 | | Bat Influenza A polymerase pre-termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
6DCF
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
6TW1
 
 | | Bat Influenza A polymerase termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2020-01-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 2020
|
|
6NPW
 
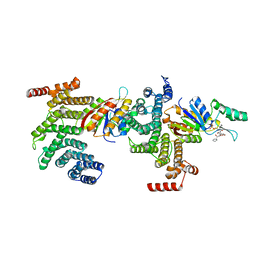 | | SSu72/Sympk in complex with Ser2/Ser5 phosphorylated peptide | | Descriptor: | PHOSPHATE ION, Ser2/Ser5 phosphorylated peptide, Ssu72 ortholog, ... | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate C-terminal domain (CTD) phosphatase during eukaryotic transcription.
J.Biol.Chem., 294, 2019
|
|
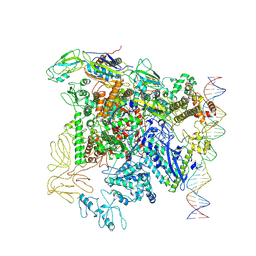
7QXI
 
 | |
6AE4
 
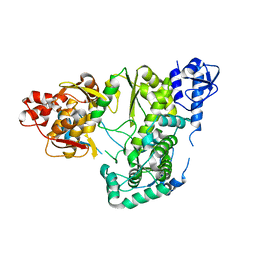 | |
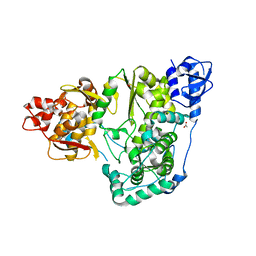
6AE6
 
 | |
6AE7
 
 | |
6AE5
 
 | |
6UTY
 
 | | E. coli sigma-S transcription initiation complex with a mismatching CTP ("Old" crystal soaked with CTP for 30 minutes) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTX
 
 | | E. coli sigma-S transcription initiation complex with an empty bubble ("Old" crystal) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UUB
 
 | | E. coli sigma-S transcription initiation complex with a mismatching UTP ("Fresh" crystal soaked with UTP for 2 hours) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.955 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UUA
 
 | | E. coli sigma-S transcription initiation complex with a mismatching CTP ("Fresh" crystal soaked with CTP for 2 hours) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
5YF5
 
 | |
4UD4
 
 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | GLYCEROL, POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.
Nucleic Acids Res., 43, 2015
|
|
4UD5
 
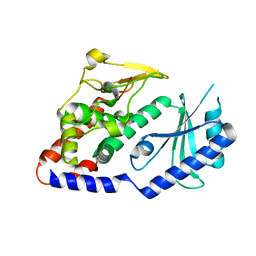 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Plasticity of Cid1 Provides a Basis for its Distributive RNA Terminal Uridylyl Transferase Activity.
Nucleic Acids Res., 43, 2015
|
|
5FMZ
 
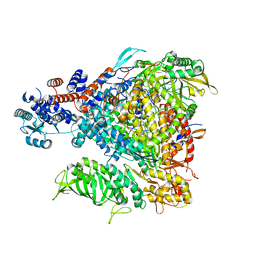 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5D4E
 
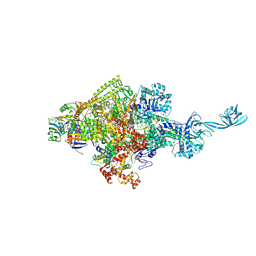 | |
6RFG
 
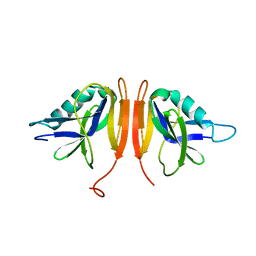 | |
7CTT
 
 | | Cryo-EM structure of Favipiravir bound to replicating polymerase complex of SARS-CoV-2 in the pre-catalytic state. | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir.
Innovation (N Y), 2, 2021
|
|
