9BB6
 
 | |
9C0Y
 
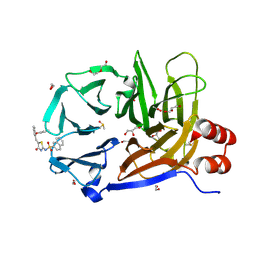 | | Clathrin terminal domain complexed with Pitstop 2c | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
9IJC
 
 | |
8XZD
 
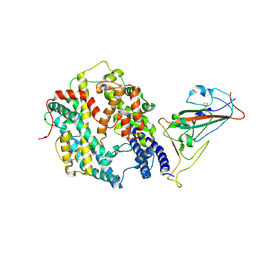 | | The structure of fox ACE2 and Omicron BF.7 RBD complex | | Descriptor: | Angiotensin-converting enzyme, Spike protein S1, ZINC ION | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-21 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 2024
|
|
9BRC
 
 | |
6DFY
 
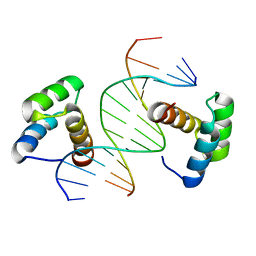 | | Remodeled crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*A)-3'), Double homeobox protein 4 | | Authors: | Aihara, H, Shi, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Comment on structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
9COZ
 
 | |
9FE7
 
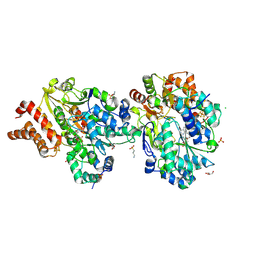 | | Crystal Structure of oxidized NuoEF variant P228R(NuoF) from Aquifex aeolicus | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8X8S
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XZ7
 
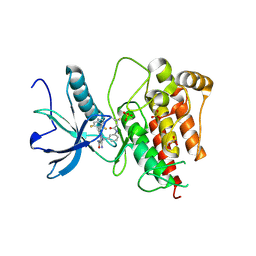 | | FGFR1 kinase domain with a covalent inhibitor 10h | | Descriptor: | 5-azanyl-3-[2-[4,6-bis(fluoranyl)-2-methyl-3~{H}-benzimidazol-5-yl]ethynyl]-1-[[3-(prop-2-enoylamino)phenyl]methyl]pyrazole-4-carboxamide, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
6DQ1
 
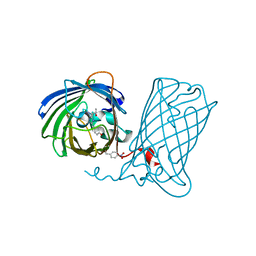 | |
8YTX
 
 | | Tubulin-RB3-TTL in complex with compound SI9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[2,3-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tubulin-RB3-TTL in complex with compound SI9
To Be Published
|
|
6DG2
 
 | |
9F14
 
 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 2024
|
|
9C5S
 
 | | Disulfide-linked, antiparallel p53-derived peptide dimer (CV1) | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Vithanage, N, Kreitler, D.K, DiGiorno, M.C, Victorio, C.G, Sawyer, N, Outlaw, V.K. | | Deposit date: | 2024-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
Res Sq, 2024
|
|
8YXP
 
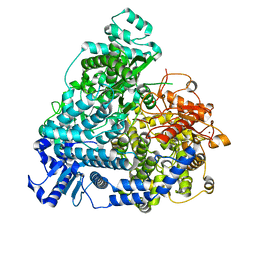 | | Structure of mumps virus L protein (state2) | | Descriptor: | RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, T.H, Shen, Q.T. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of the mumps virus polymerase complex via cryo-electron microscopy.
Nat Commun, 15, 2024
|
|
9EOU
 
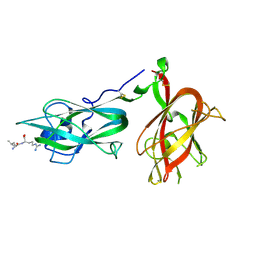 | |
9BB5
 
 | |
9EX0
 
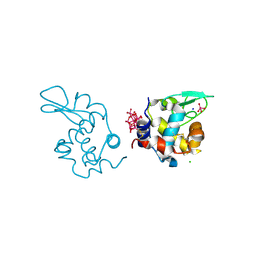 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure A) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8ZNE
 
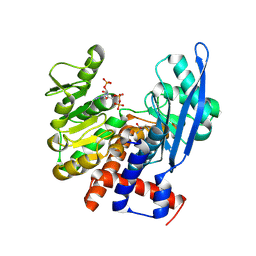 | |
8ZN4
 
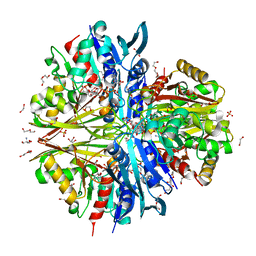 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
6DH9
 
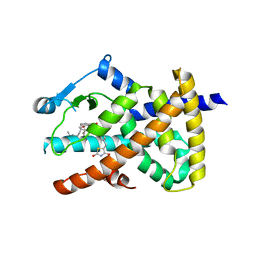 | |
9EX2
 
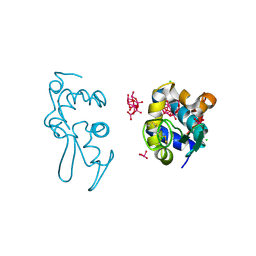 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure C) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9C11
 
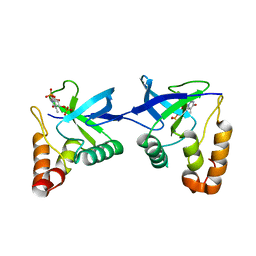 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-swapping promoted by the introduction of a charge in the hydrophobic interior of a protein
To Be Published
|
|
8ZKD
 
 | | The Crystal Structure of the RON from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Macrophage-stimulating protein receptor beta chain, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Pan, W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the RON from Biortus.
To Be Published
|
|
