5GYR
 
 | | Tetrameric Allochromatium vinosum cytochrome c' | | Descriptor: | Cytochrome c', HEME C | | Authors: | Yamanaka, M, Hoshizumi, M, Nagao, S, Nakayama, R, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2016-09-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation and carbon monoxide-dependent dissociation of Allochromatium vinosum cytochrome c' oligomers using domain-swapped dimers
Protein Sci., 26, 2017
|
|
8CVD
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVF
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVH
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVA
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CV9
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVG
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CV8
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
2KM4
 
 | | Solution structure of Rtt103 CTD interacting domain | | Descriptor: | Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S, Kim, M, Leeper, T.C, Becker, R, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2009-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2DAH
 
 | | Solution Structure of the C-terminal UBA Domain in the Human Ubiquilin 3 | | Descriptor: | Ubiquilin-3 | | Authors: | Zhao, C, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal UBA Domain in the Human Ubiquilin 3
To be Published
|
|
9BIF
 
 | | Fab B11-OspCA complex | | Descriptor: | CHLORIDE ION, Outer surface protein C, PRASEODYMIUM ION, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure of a Human Monoclonal Antibody in Complex with Outer Surface Protein C of the Lyme Disease Spirochete, Borreliella burgdorferi.
J Immunol., 213, 2024
|
|
2KCZ
 
 | | Solution NMR structure of the C-terminal domain of protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D | | Descriptor: | Uncharacterized protein DR_A0006 | | Authors: | Mills, J.L, Ghosh, A, Garcia, E, Wang, H, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the C-Terminal Domain of Protein DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D.
To be Published
|
|
8RDE
 
 | |
2KRH
 
 | |
6ISJ
 
 | | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans
To be published
|
|
1MC7
 
 | | Solution Structure of mDvl1 PDZ domain | | Descriptor: | Segment polarity protein dishevelled homolog DVL-1 | | Authors: | Wong, H.-C, Bourdelas, A, Shao, Y, Wu, D, Shi, D.L, Zheng, J. | | Deposit date: | 2002-08-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled
Mol.Cell, 12, 2003
|
|
6FF5
 
 | |
6CZQ
 
 | |
3HPX
 
 | |
4ZRL
 
 | | Structure of the non canonical Poly(A) polymerase complex GLD-2 - GLD-3 | | Descriptor: | CHLORIDE ION, Defective in germ line development protein 3, Poly(A) RNA polymerase gld-2 | | Authors: | Nakel, K, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2015-05-12 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KJQ
 
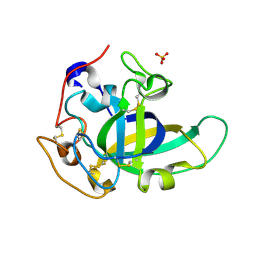 | | X-ray structure of PcCel45A in complex with cellobiose expressed in Aspergillus nidullans | | Descriptor: | Endoglucanase V-like protein, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Godoy, A.S, Ramia, M.P, Camilo, C.M, Polikarpov, I. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structure, computational and biochemical analysis of PcCel45A endoglucanase from Phanerochaete chrysosporium and catalytic mechanisms of GH45 subfamily C members.
Sci Rep, 8, 2018
|
|
4XMO
 
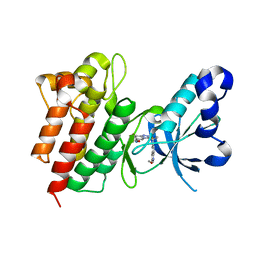 | | Crystal structure of c-Met in complex with (R)-5-(8-fluoro-3-(1-fluoro-1-(3-methoxyquinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1R)-1-fluoro-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-methoxyquinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
