7EXK
 
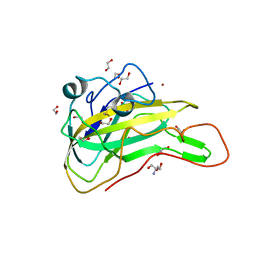 | | An AA9 LPMO of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
7EQ1
 
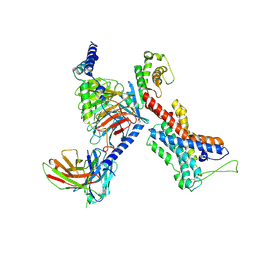 | | GPR114-Gs-scFv16 complex | | Descriptor: | Adhesion G-protein coupled receptor G5, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
4L4W
 
 | |
4NBC
 
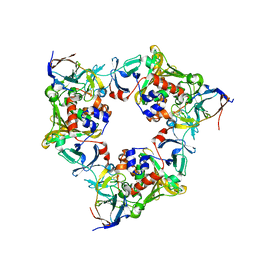 | | Oxygenase with Phe275 replaced by Trp and ferredoxin complex of carbazole 1,9a-dioxygenase (form1) | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4HDQ
 
 | |
4GWS
 
 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase with Filled Central Cavity | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Gao, Y, Honzatko, R.B. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hydrophobic Central Cavity in Fructose-1,6-bisphosphatase is Essential for the Synergism in AMP/Fructose 2,6-bisphosphate Inhibition
To be Published
|
|
4GWU
 
 | | Crystal Structure of Fru 2,6-bisphosphate complexes of Porcine Liver Fructose-1,6-bisphosphatase with Filled Central Cavity | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION | | Authors: | Gao, Y, Honzatko, R.B. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hydrophobic Central Cavity in Fructose-1,6-bisphosphatase is Essential for the Synergism in AMP/Fru 2,6-bisphosphate Inhibition
To be Published
|
|
5UKM
 
 | | bovine GRK2 in complex with human Gbetagamma subunits and CCG258208 (14as) | | Descriptor: | 5-[(3S,4R)-3-{[(2H-1,3-benzodioxol-5-yl)oxy]methyl}piperidin-4-yl]-2-fluoro-N-[(1H-pyrazol-5-yl)methyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cruz-Rodriguez, O, Tesmer, J.J.G. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Based Design of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors Based on Paroxetine.
J. Med. Chem., 60, 2017
|
|
7E29
 
 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Smolle, M, Liang, H, Liu, Y. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
7E2P
 
 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2Q
 
 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
4JAN
 
 | | crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
5UKK
 
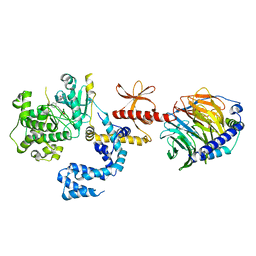 | | Human GRK2 in complex with human G-beta-gamma subunits and CCG211998 (14ak) | | Descriptor: | 5-[(3S,4R)-3-{[(2H-1,3-benzodioxol-5-yl)oxy]methyl}piperidin-4-yl]-2-fluoro-N-[(pyridin-2-yl)methyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cato, M.C, Homan, K.T, Tesmer, J.J.G. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors Based on Paroxetine.
J. Med. Chem., 60, 2017
|
|
4N9N
 
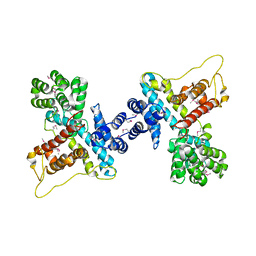 | |
4KAM
 
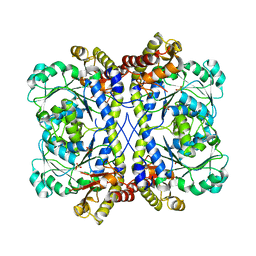 | |
5UKL
 
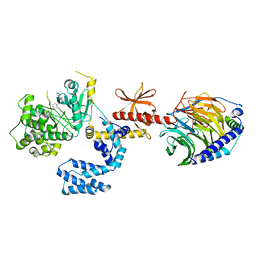 | | Human GRK2 in complex with Gbetagamma subunits and CCG222886 (14bd) | | Descriptor: | 2-{5-[(3S,4R)-3-{[(2H-1,3-benzodioxol-5-yl)oxy]methyl}piperidin-4-yl]-2-fluorophenyl}-N-[2-(1H-pyrazol-4-yl)ethyl]acetamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cato, M.C, Homan, K.T, Tesmer, J.J.G. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Highly Selective and Potent G Protein-Coupled Receptor Kinase 2 Inhibitors Based on Paroxetine.
J. Med. Chem., 60, 2017
|
|
4JAM
 
 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | Descriptor: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
7DMB
 
 | |
7JHA
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JI7
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J33 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*CP*AP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JHV
 
 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J36 immobile Holliday junction with R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.058 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JI6
 
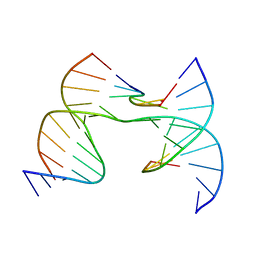 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble junction version) containing the J34 immobile Holliday junction with R3 symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*GP*TP*TP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKE
 
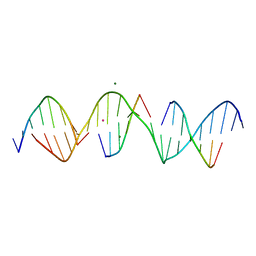 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J2 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JKD
 
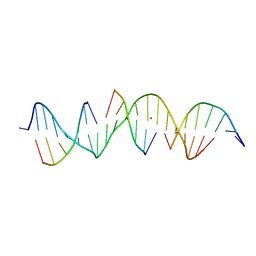 | | Self-assembly of a 3D DNA crystal lattice (4x6 scramble duplex version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*AP*CP*GP*AP*CP*AP*CP*TP*GP*AP*CP*GP*GP*AP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*CP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.058 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
7JFW
 
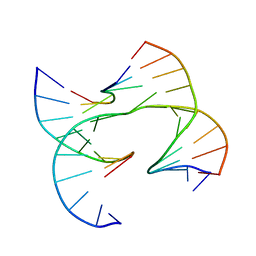 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J10 immobile Holliday junction | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*TP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*CP*AP*CP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.012 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
