4EHQ
 
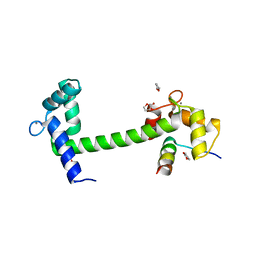 | | Crystal Structure of Calmodulin Binding Domain of Orai1 in Complex with Ca2+/Calmodulin Displays a Unique Binding Mode | | Descriptor: | CALCIUM ION, Calcium release-activated calcium channel protein 1, Calmodulin, ... | | Authors: | Liu, Y, Zheng, X, Mueller, G.A, Sobhany, M, DeRose, E.F, Zhang, Y, London, R.E, Birnbaumer, L. | | Deposit date: | 2012-04-03 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9005 Å) | | Cite: | Crystal structure of calmodulin binding domain of orai1 in complex with ca2+*calmodulin displays a unique binding mode.
J.Biol.Chem., 287, 2012
|
|
4E7E
 
 | |
6E80
 
 | |
6E84
 
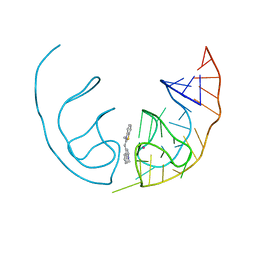 | | Crystal structure of the Corn aptamer in complex with TO | | Descriptor: | 1-methyl-4-[(Z)-(3-methyl-1,3-benzothiazol-2(3H)-ylidene)methyl]quinolin-1-ium, POTASSIUM ION, RNA (36-MER) | | Authors: | Sjekloca, L, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Binding between G Quadruplexes at the Homodimer Interface of the Corn RNA Aptamer Strongly Activates Thioflavin T Fluorescence.
Cell Chem Biol, 26, 2019
|
|
4EOH
 
 | | Crystal Structure of Human PL Kinase with bound Theophylline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Pyridoxal Kinase, SODIUM ION, ... | | Authors: | Safo, M.K, Gandhi, A.K, Musayev, F.N. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human pyridoxal kinase in complex with the neurotoxins, ginkgotoxin and theophylline: insights into pyridoxal kinase inhibition.
Plos One, 7, 2012
|
|
6E82
 
 | |
2PNS
 
 | | 1.9 Angstrom resolution crystal structure of a plant cysteine protease Ervatamin-C refinement with cDNA derived amino acid sequence | | Descriptor: | Ervatamin-C, a papain-like plant cysteine protease, PHOSPHATE ION, ... | | Authors: | Ghosh, R, Guha Thakurta, P, Biswas, S, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A thermostable cysteine protease precursor from a tropical plant contains an unusual C-terminal propeptide: cDNA cloning, sequence comparison and molecular modeling studies.
Biochem.Biophys.Res.Commun., 362, 2007
|
|
1ZO3
 
 | | The P-site and P/E-site tRNA structures fitted to P/I site codon. | | Descriptor: | tRNA | | Authors: | Allen, G.S, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13.8 Å) | | Cite: | The Cryo-EM Structure of a Translation Initiation Complex from Escherichia coli.
Cell(Cambridge,Mass.), 121, 2005
|
|
5W0J
 
 | |
4EOF
 
 | | Lysozyme in the presence of arginine | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-04-14 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation.
Sci Rep, 6, 2016
|
|
1ZJ5
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZIC
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S, R206A) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZO2
 
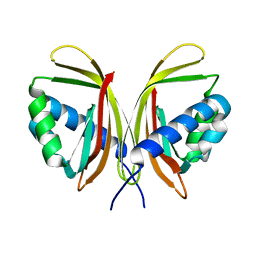 | | Structure of nuclear transport factor 2 (Ntf2) from Cryptosporidium parvum | | Descriptor: | nuclear transport factor 2 | | Authors: | Choe, J, Artz, J.D, Gao, M, Lew, J, Zhao, Y, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
7MHV
 
 | |
1YHC
 
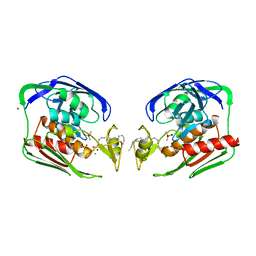 | | Crystal structure of Aquifex aeolicus LpxC deacetylase complexed with cacodylate | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hernick, M, Gennadios, H.A, Whittington, D.A, Rusche, K.M, Christianson, D.W, Fierke, C.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase Functions through a General Acid-Base Catalyst Pair Mechanism
J.Biol.Chem., 280, 2005
|
|
1YF4
 
 | |
6G67
 
 | |
6G6A
 
 | |
6G6D
 
 | |
6G6G
 
 | |
4DFU
 
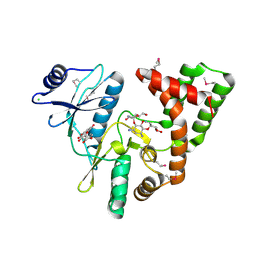 | | Inhibition of an antibiotic resistance enzyme: crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA in complex with kanamycin inhibited with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, APH(2")-Id, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4DEB
 
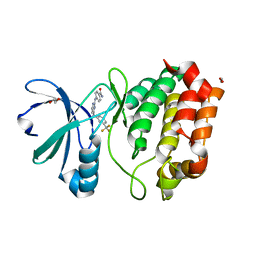 | | Aurora A in complex with RK2-17-01 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[3-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzamide, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
4DIL
 
 | | Flavo Di-iron protein H90N mutant from Thermotoga maritima | | Descriptor: | CHLORIDE ION, FLAVOPROTEIN, MU-OXO-DIIRON | | Authors: | Fang, H, Caranto, J.D, Taylor, A.B, Hart, P.J, Kurtz, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Histidine ligand variants of a flavo-diiron protein: effects on structure and activities.
J.Biol.Inorg.Chem., 17, 2012
|
|
1Y7T
 
 | | Crystal structure of NAD(H)-depenent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
