1TKB
 
 | |
1TKC
 
 | |
1TKA
 
 | |
6BQB
 
 | | MGG4 Fab in complex with peptide | | Descriptor: | GLYCEROL, MGG4 Fab heavy chain, MGG4 Fab light chain, ... | | Authors: | Oyen, D, Tan, J, Lanzavecchia, A, Wilson, I.A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-07 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein.
Nat. Med., 24, 2018
|
|
6W78
 
 | | crystal structure of a plant ice-binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antifreeze polypeptide | | Authors: | Wang, Y.N, Zhang, H.Q. | | Deposit date: | 2020-03-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Carrot 'antifreeze' protein has an irregular ice-binding site that confers weak freezing point depression but strong inhibition of ice recrystallization.
Biochem.J., 477, 2020
|
|
6W7D
 
 | | K2P2.1 (TREK-1), 10 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6W8L
 
 | | Crystal structure of JAK1 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
5ZDR
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ascofuranone derivative | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5ZEC
 
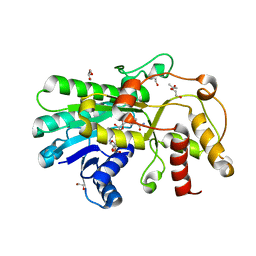 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
6VKC
 
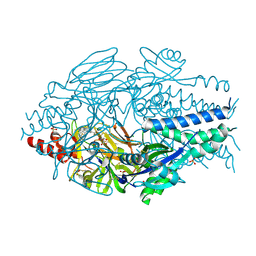 | | Crystal Structure of Inhibitor JNJ-36811054 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 3-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-imidazo[4,5-b]pyridin-2-yl]methyl}-1-cyclopropyl-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
5ZDQ
 
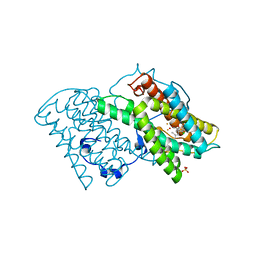 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with COLLETOCHLORIN B | | Descriptor: | 3-chloro-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-4,6-dihydroxy-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
7LXT
 
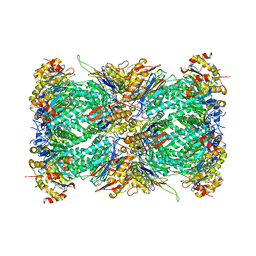 | | Structure of Plasmodium falciparum 20S proteasome with bound bortezomib | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Morton, C.J, Metcalfe, R.D, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXU
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound MPI-5 | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Xie, S.C, Liu, B, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4A7I
 
 | | Factor Xa in complex with a potent 2-amino-ethane sulfonamide inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID [2-(1--ISOPROPYL-PIPERIDIN-4-YLSULFAMOYL)-ETHYL]-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN XA, CALCIUM ION, ... | | Authors: | Nazare, M, Matter, H, Will, D.W, Wagner, M, Urmann, M, Czech, J, Schreuder, H, Bauer, A, Ritter, K, Wehner, V. | | Deposit date: | 2011-11-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment Deconstruction of Small, Potent Factor Xa Inhibitors: Exploring the Superadditivity Energetics of Fragment Linking in Protein-Ligand Complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7LXV
 
 | | Structure of human 20S proteasome with bound MPI-5 | | Descriptor: | N-[(1R)-2-([1,1'-biphenyl]-4-yl)-1-boronoethyl]-1-methyl-L-prolinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6UT0
 
 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
4ALZ
 
 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
6CQS
 
 | | Sediminispirochaeta smaragdinae SPS-1 metallo-beta-lactamase | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Page, R.C, VanPelt, J, Cheng, Z, Crowder, M.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Metal Center Drives the Activity of the Sediminispirochaeta smaragdinae Metallo-beta-lactamase SPS-1.
Biochemistry, 57, 2018
|
|
6O21
 
 | | Crystal Structure of Human KLK4 in Complex With Cleaved SFTI-FCQR(Asn14)[1,14] Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Kallikrein 4 (Prostase, enamel matrix, ... | | Authors: | Ilyichova, O.V, Buckle, A.M. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
6NZA
 
 | |
3URR
 
 | |
