4JGH
 
 | | Structure of the SOCS2-Elongin BC complex bound to an N-terminal fragment of Cullin5 | | Descriptor: | Cullin-5, Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, ... | | Authors: | Kim, Y.K, Kwak, M.J, Ku, B, Suh, H.Y, Joo, K, Lee, J, Jung, J.U, Oh, B.H. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HA2
 
 | | Crystal structure ofa phenyl alanine 91 mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION | | Authors: | Majumder, S, Sen, U. | | Deposit date: | 2012-09-25 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved tryptophan (W91) at the barrel-lid junction modulates the packing and stability of Kunitz (STI) family of inhibitors.
Biochim.Biophys.Acta, 1854, 2015
|
|
4H9W
 
 | | crystal structure of a METHIONINE mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Majumder, S, Sen, U. | | Deposit date: | 2012-09-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved tryptophan (W91) at the barrel-lid junction modulates the packing and stability of Kunitz (STI) family of inhibitors.
Biochim.Biophys.Acta, 1854, 2015
|
|
4GQB
 
 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4J3Q
 
 | | Crystal structure of truncated catechol oxidase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Hakulinen, N, Gasparetti, C, Kaljunen, H, Rouvinen, J. | | Deposit date: | 2013-02-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae.
J.Biol.Inorg.Chem., 18, 2013
|
|
4J3P
 
 | | Crystal structure of full-length catechol oxidase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Hakulinen, N, Gasparetti, C, Kaljunen, H, Rouvinen, J. | | Deposit date: | 2013-02-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae.
J.Biol.Inorg.Chem., 18, 2013
|
|
2GWA
 
 | | Crystal Structure of a Complex Formed Between the DNA Holliday Junction and a Bis-Acridine Molecule. | | Descriptor: | 5'-D(*Tp*Cp*Gp*Gp*Tp*Ap*Cp*Cp*Gp*A)-3', 9,9'-(HEXANE-1,6-DIYLDIIMINO)BIS{N-[2-(DIMETHYLAMINO)ETHYL]ACRIDINE-4-CARBOXAMIDE}, SPERMINE | | Authors: | Brogden, A.L, Hopcroft, N.H, Cardin, C.J, Searcey, M. | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ligand bridging of the DNA Holliday junction: molecular recognition of a stacked-X four-way junction by a small molecule.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
4HQB
 
 | |
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4M3P
 
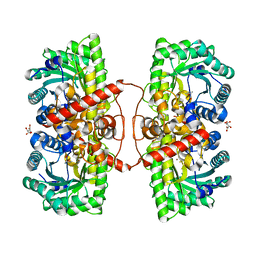 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
4LFD
 
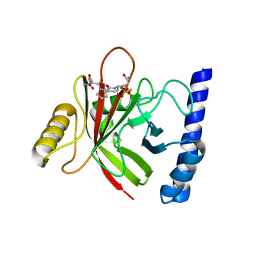 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
4N72
 
 | |
4NOA
 
 | |
4MBQ
 
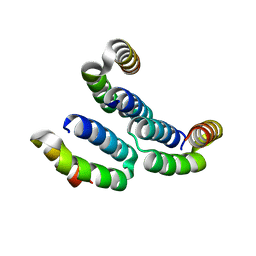 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4M6R
 
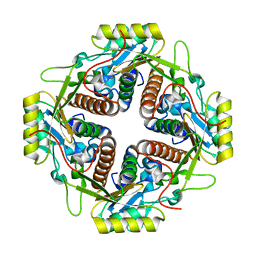 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NPH
 
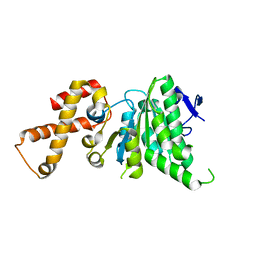 | | Crystal structure of SsaN from Salmonella enterica | | Descriptor: | Probable secretion system apparatus ATP synthase SsaN | | Authors: | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
6WED
 
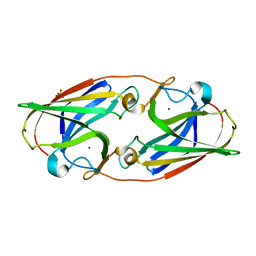 | |
6WEE
 
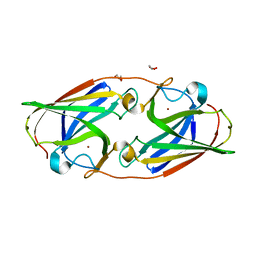 | | Copper-bound M88I variant of Campylobacter jejuni P19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2020-04-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A copper site is required for iron transport by the periplasmic proteins P19 and FetP.
Metallomics, 12, 2020
|
|
6WEF
 
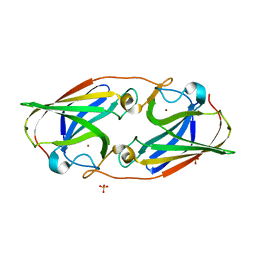 | |
4P1Q
 
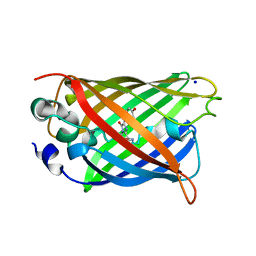 | | GREEN FLUORESCENT PROTEIN E222H VARIANT | | Descriptor: | Green fluorescent protein, SODIUM ION | | Authors: | Klein, M, Carius, Y, Auerbach, D, Franz, S, Jung, G, Lancaster, C.R.D. | | Deposit date: | 2014-02-27 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Replacement of Highly Conserved E222 by the Photostable Non-photoconvertible Histidine in GFP.
Chembiochem, 15, 2014
|
|
4NOE
 
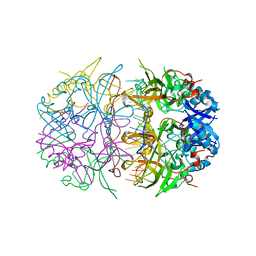 | | Crystal structure of DdrB bound to 30b ssDNA | | Descriptor: | 5'-D(*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP)-3', CALCIUM ION, Single-stranded DNA-binding protein DdrB | | Authors: | Sugiman-Marangos, S.N, Weiss, Y.M, Junop, M.S. | | Deposit date: | 2013-11-19 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9B3P
 
 | |
2IXZ
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*CP*UP*GP*UP*GP*CP*CP)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2IXY
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
7P1K
 
 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
