6VF5
 
 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (120 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (120 min)
To be published
|
|
6VF9
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Ternary Complex, 50 mM Mg2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Ternary Complex, 50 mM Mg2+ (2160 min)
To be published
|
|
6VF7
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | DNA Polymerase Mu, 8-oxodGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min)
To be published
|
|
6VFA
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Ground State Ternary Complex, 50 mM Mn2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Ground State Ternary Complex, 50 mM Mn2+ (15 min)
To be published
|
|
6VFB
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Reaction State Ternary Complex, 50 mM Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Reaction State Ternary Complex, 50 mM Mn2+ (960 min)
To be published
|
|
6VFC
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min)
To be published
|
|
161D
 
 | |
6WGG
 
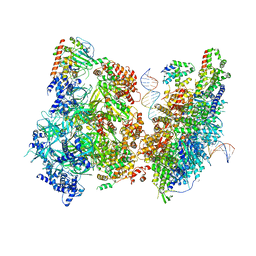 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4YJ0
 
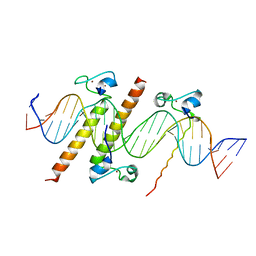 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | Descriptor: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | Authors: | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5JVW
 
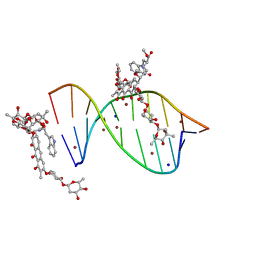 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
189D
 
 | |
188D
 
 | |
1AKZ
 
 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4QTJ
 
 | | Complex of WOPR domain of Wor1 in Candida albicans with the 13bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
6GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
6O6K
 
 | | HUaa 19bp SYM DNA pH 5.5 | | Descriptor: | DNA (5'-D(P*AP*TP*TP*TP*CP*AP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*AP*TP*CP*AP*TP*GP*AP*AP*A)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
6WGC
 
 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OAJ
 
 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
3G73
 
 | | Structure of the FOXM1 DNA binding | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(P*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein M1, ... | | Authors: | Littler, D.R, Perrakis, A, Hibbert, R.G, Medema, R.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence
Nucleic Acids Res., 2010
|
|
6O8Q
 
 | | HUaa 19bp SYM DNA pH 4.5 | | Descriptor: | DNA (57-MER), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.216 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | Descriptor: | DNA (44-MER), DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
3F2B
 
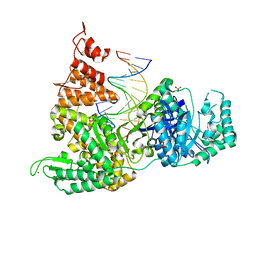 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mg and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3TEK
 
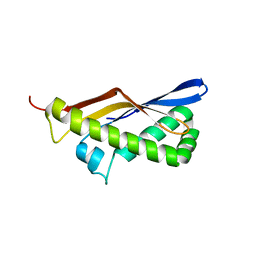 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1XHX
 
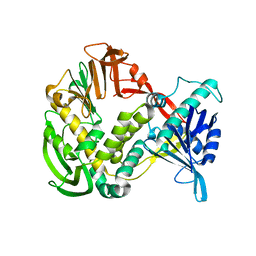 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
