7P7M
 
 | | Complex I from E. coli, DDM/LMNG-purified, inhibited by Piericidin A, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
8BHB
 
 | | GABA-A receptor a5 homomer - a5V3 - RO154513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, ethyl 8-[(azanylidene-$l^{4}-azanylidene)amino]-5-methyl-6-oxidanylidene-4~{H}-imidazo[1,5-a][1,4]benzodiazepine-3-carboxylate | | Authors: | Miller, P.S, Malinauskas, T.M, Hardwick, S.W, Chirgadze, D.Y, Wahid, A.A. | | Deposit date: | 2022-10-30 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BGI
 
 | | GABA-A receptor a5 homomer - a5V1 - Flumazenil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-5, Pregnanolone, ... | | Authors: | Miller, P.S, Malinauskas, T.M, Omari, K.E, Aricescu, A.R. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The molecular basis of drug selectivity for alpha 5 subunit-containing GABA A receptors.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6ZNU
 
 | | MaeB PTA domain E544R mutant | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6ZNT
 
 | | MaeB PTA domain, acetyl-CoA bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
8C0Z
 
 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
6FQM
 
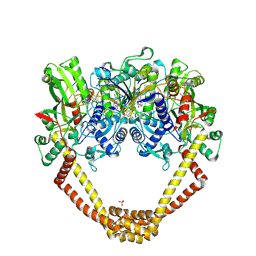 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | Descriptor: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
9DIQ
 
 | | Crystal structure of Apo-H5 hemagglutinin from the influenza virus A/Texas/37/2024 (H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A single mutation in bovine influenza H5N1 hemagglutinin switches specificity to human receptors.
Science, 386, 2024
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
7R6R
 
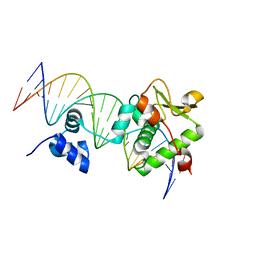 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
6FBF
 
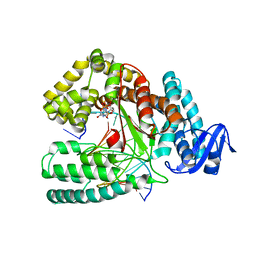 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fourth primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*GP*GP*TP*GP*CP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*AP*(OH3)P*CP*GP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FCQ
 
 | |
6FBD
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the second primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*AP*(OH3)P*C)-3'), DNA (5'-D(P*AP*AP*AP*CP*GP*GP*TP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBH
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the sixth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*GP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW1
 
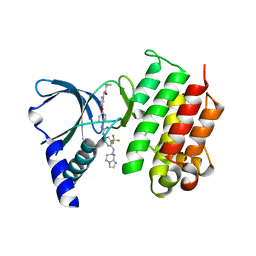 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
6GIW
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-a | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
6ZVJ
 
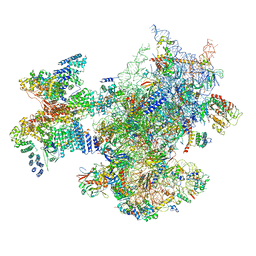 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7A09
 
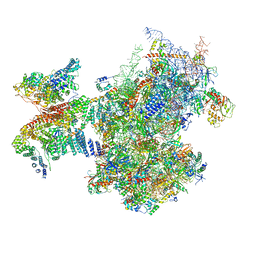 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State III | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
8FYN
 
 | | MicroED structure of A2A from plasma milled lamellae | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
8CLG
 
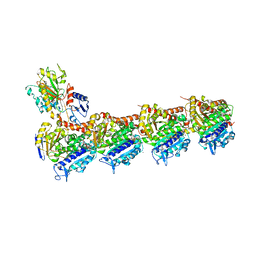 | | Epothilone A and Colchicine bound to tubulin (T2R-TTL) complex | | Descriptor: | CALCIUM ION, EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
6FBC
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification at the 3'-terminus of the primer. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBG
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fifth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*CP*GP*GP*TP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
