8SOJ
 
 | |
8SOK
 
 | |
8OTS
 
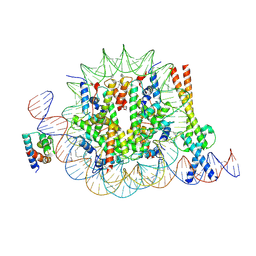 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSI
 
 | | Genetically encoded green ratiometric calcium indicator FNCaMP in calcium-bound state | | Descriptor: | CALCIUM ION, mNeonGreen,Calmodulin,Contig An16c0100, genomic contig | | Authors: | Varfolomeeva, L.A, Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FNCaMP, ratiometric green calcium indicator based on mNeonGreen protein.
Biochem.Biophys.Res.Commun., 665, 2023
|
|
8OVN
 
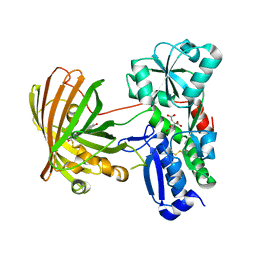 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OVO
 
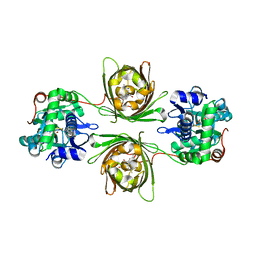 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVP
 
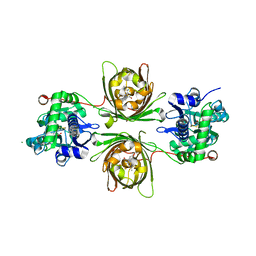 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
7YDQ
 
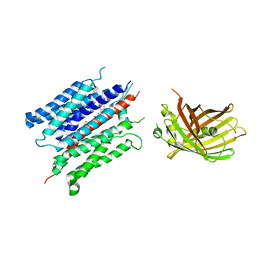 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
8FCK
 
 | | Structure of the vertebrate augmin complex | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | Authors: | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.88 Å) | | Cite: | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
8F6P
 
 | | Rat Cardiac Sodium Channel with Ranolazine Bound | | Descriptor: | (R)-ranolazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lenaeus, M.J, Tonggu, L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the cardiac sodium channel by the atypical antiarrhythmic drug ranolazine.
Nat Cardiovasc Res, 2, 2023
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
8BVG
 
 | |
7ZCT
 
 | |
8B65
 
 | | Structure of rsCherry crystallized in anaerobic conditions | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxygen-induced chromophore degradation in the photoswitchable red fluorescent protein rsCherry.
Int.J.Biol.Macromol., 239, 2023
|
|
8B7G
 
 | | Structure of oxygen-degraded rsCherry | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxygen-induced chromophore degradation in the photoswitchable red fluorescent protein rsCherry.
Int.J.Biol.Macromol., 239, 2023
|
|
8C0N
 
 | | Crystal structure of the red form of the mTagFT fluorescent timer | | Descriptor: | Blue-to-red TagFT fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Agapova, Y.K, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blue-to-Red TagFT, mTagFT, mTsFT, and Green-to-FarRed mNeptusFT2 Proteins, Genetically Encoded True and Tandem Fluorescent Timers.
Int J Mol Sci, 24, 2023
|
|
8FED
 
 | | Structure of Mce1-LucB complex from Mycobacterium smegmatis (Map1) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEE
 
 | | Structure of Mce1 transporter from Mycobacterium smegmatis in the absence of LucB (Map2) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8FEF
 
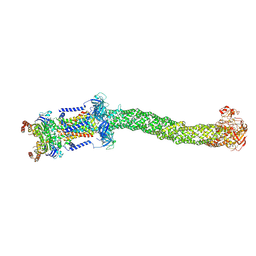 | | Structure of Mce1 transporter from Mycobacterium smegmatis (Map0) | | Descriptor: | ABC transporter, ATP-binding protein,Green fluorescent protein chimera, ABC-transporter integral membrane protein, ... | | Authors: | Chen, J, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of an endogenous mycobacterial MCE lipid transporter.
Nature, 620, 2023
|
|
8BGL
 
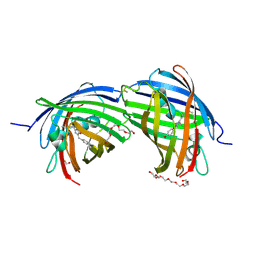 | | Structure of the dimeric rsCherryRev1.4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PAmCherry1 protein, ... | | Authors: | Bui, T.Y.H, Van Meervelt, L. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An unusual disulfide-linked dimerization in the fluorescent protein rsCherryRev1.4.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8ET3
 
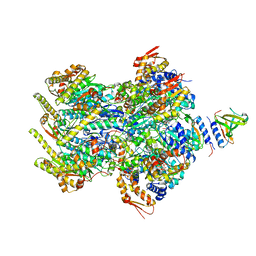 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TSV
 
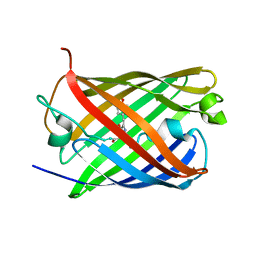 | | Room temperature rsEospa Trans-state structure at pH 5.5 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSU
 
 | | Room temperature rsEospa Cis-state structure at pH 5.5 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSS
 
 | | Room temperature rsEospa Trans-state structure at pH 8.4 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSR
 
 | | Room temperature rsEospa Cis-state structure at pH 8.4 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
