6OFH
 
 | |
8ELM
 
 | | Apo human biliverdin reductase beta (293K) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
8ELL
 
 | | Apo human biliverdin reductase beta (cryogenic) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
8BYX
 
 | |
4XPU
 
 | | The crystal structure of EndoV from E.coli | | Descriptor: | Endonuclease V | | Authors: | Xie, W, Zhang, Z. | | Deposit date: | 2015-01-18 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of E. coli endonuclease V, an essential enzyme for deamination repair
Sci Rep, 5, 2015
|
|
6OFF
 
 | |
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
6NCU
 
 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8BZM
 
 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 2025
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
9BYK
 
 | |
4U7E
 
 | | The crystal structure of the complex of LIP5 NTD and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
4U7Y
 
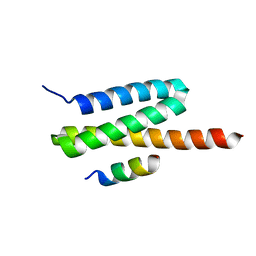 | | Structure of the complex of VPS4B MIT and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein 4B | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
4U7I
 
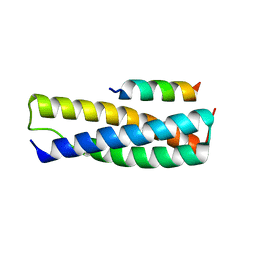 | | Structure of the complex of Spartin MIT and IST1 MIM | | Descriptor: | IST1 homolog, Spartin | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
7RYE
 
 | |
8CXH
 
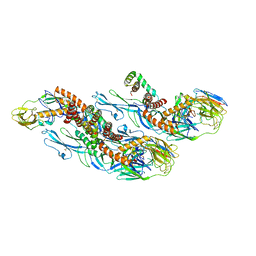 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C10 heavy chain, C10 light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXI
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXG
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C8 scFv heavy chain, C8 scFv light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8JJ6
 
 | | Structure of the NELF-BCE complex | | Descriptor: | NELF-E, Negative elongation factor B, Negative elongation factor complex member C/D | | Authors: | Wang, Z, Cao, Y, Qin, Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of the human negative elongation factor NELF-B/C/E ternary complex.
Biochem.Biophys.Res.Commun., 677, 2023
|
|
8R7Z
 
 | | transcription factor BARHL2 homodimer with spacing four bp | | Descriptor: | BarH-like 2 homeobox protein, DNA (5'-D(P*AP*CP*CP*GP*TP*TP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*AP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 641, 2025
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5NO6
 
 | | TEAD4-HOXB13 complex bound to DNA | | Descriptor: | DNA, Homeobox protein Hox-B13, Transcriptional enhancer factor TEF-3 | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2017-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 2025
|
|
9B0W
 
 | |
9AZN
 
 | |
