8ANW
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8). | | Descriptor: | Capsid protein, VP0, VP1, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris .
Viruses, 14, 2022
|
|
4ZU4
 
 | | X-ray structure of the 3,4-ketoisomerase domain of FdtD from Shewanella denitrificans | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
5F7P
 
 | |
5J4V
 
 | | The crystal structure of Inhibitor Bound to JCV Helicase | | Descriptor: | 2-(2-phenoxypyridin-3-yl)[1,3]thiazolo[5,4-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Large T antigen, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5J47
 
 | | The X-ray structure of Inhibitor Bound to JCV Helicase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-phenoxyphenyl)[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole, Large T antigen, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
1H3G
 
 | | Cyclomaltodextrinase from Flavobacterium sp. No. 92: from DNA sequence to protein structure | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase | | Authors: | Fritzsche, H.B, Schwede, T, Jelakovic, S, Schulz, G.E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-08-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Covalent and Three-Dimensional Structure of the Cyclodextrinase from Flavobacterium Sp. No. 92.
Eur.J.Biochem., 270, 2003
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5FAN
 
 | | Crystal structure of PvHCT in complex with p-coumaroyl-CoA and protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase 2, p-coumaroyl-CoA | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
6NJ7
 
 | | 11-BETA DEHYDROGENASE ISOZYME 1 IN COMPLEX WITH COLLETOIC ACID | | Descriptor: | (1S,4S,5S,9S)-9-hydroxy-8-methyl-4-(propan-2-yl)spiro[4.5]dec-7-ene-1-carboxylic acid, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Miller, D.J, Rivas, F. | | Deposit date: | 2019-01-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic Insight on the Mode of Action of Colletoic Acid.
J.Med.Chem., 62, 2019
|
|
1H2B
 
 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
5FCL
 
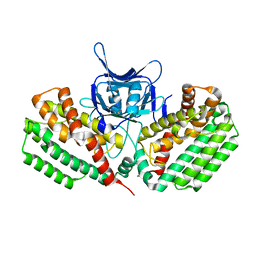 | | Crystal structure of Cas1 from Pectobacterium atrosepticum | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Wilkinson, M.E, Nakatani, Y, Opel-Reading, H.K, Fineran, P.C, Krause, K.L. | | Deposit date: | 2015-12-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and in vivo activity of Cas1 from the type I-F CRISPR-Cas system.
Biochem.J., 473, 2016
|
|
6D1O
 
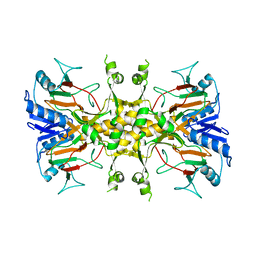 | | FT_5 dioxygenase apoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
1H50
 
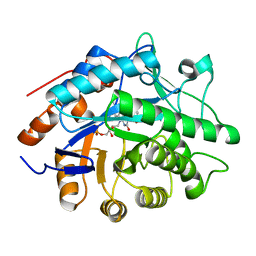 | | Structure of Pentaerythritol Tetranitrate Reductase and complexes | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2001-05-17 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
5X1F
 
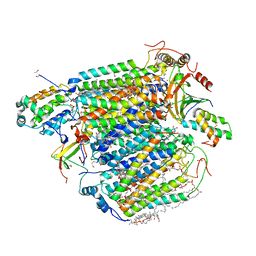 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
1AIZ
 
 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, CADMIUM ION, SULFATE ION | | Authors: | Baker, E.N, Anderson, B.F, Blackwell, K.A. | | Deposit date: | 1993-11-11 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6D31
 
 | | Structure of human Usb1 with adenosine 5'-monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nomura, Y, Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2018-04-14 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and mechanistic basis for preferential deadenylation of U6 snRNA by Usb1.
Nucleic Acids Res., 46, 2018
|
|
1HCR
 
 | |
3HRI
 
 | |
1H8Y
 
 | | Crystal structure of the class D beta-lactamase OXA-13 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BETA-LACTAMASE, SULFATE ION | | Authors: | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | Deposit date: | 2001-02-17 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
5UIP
 
 | | structure of DHFR with bound DAP, p-ABG and NADP | | Descriptor: | Dihydrofolate reductase, N-(4-aminobenzene-1-carbonyl)-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Basis for Biguanide Activity.
Biochemistry, 56, 2017
|
|
6NTA
 
 | | Modified ASL proline bound to Thermus thermophilus 70S (cognate) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hoffer, E.D, Maehigashi, T, Subaramanian, S, Hong, S, Dunham, C.M. | | Deposit date: | 2019-01-28 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6WBO
 
 | |
6NSY
 
 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.263 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6CD7
 
 | | Crystal structure of APH(2")-IVa in complex with plazomicin | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, APH(2'')-Id, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Di Leo, R, Savchenko, A, Satchell, K.J, Joachimiak, J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5A5P
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 8-2-(dimethylamino)ethylamino-3-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(dimethylamino)ethyl]amino}-3-methyl-1,2-dihydroquinolin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
