6G2H
 
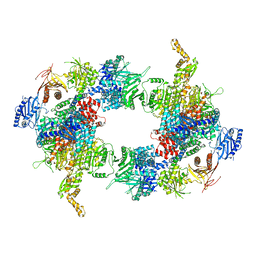 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
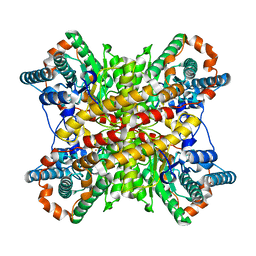
2XIN
 
 | |
4A6Q
 
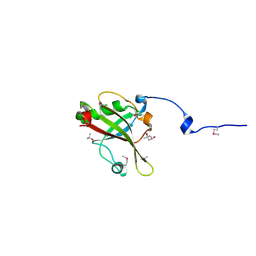 | | Crystal structure of mouse SAP18 residues 6-143 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, ISOPROPYL ALCOHOL | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-08 | | Release date: | 2012-03-07 | | Last modified: | 2015-03-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
7KLP
 
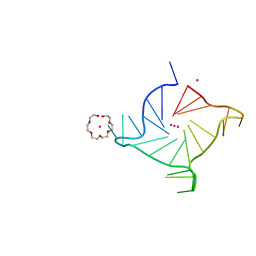 | | Crystal structure of a three-tetrad, parallel, K+ stabilized human telomeric G-quadruplex | | Descriptor: | HEXAETHYLENE GLYCOL, POTASSIUM ION, telomeric DNA | | Authors: | Yatsunyk, L.A, Li, K.S, Chen, E.V. | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Water spines and networks in G-quadruplex structures.
Nucleic Acids Res., 49, 2021
|
|
4A8X
 
 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ZI5
 
 | | Crystal Structure of Dienelactone Hydrolase-like Promiscuous Phospotriesterase P91 from Metagenomic Libraries | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, P91 | | Authors: | Colin, P.-Y, Fischer, G, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2015-04-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics.
Nat Commun, 6, 2015
|
|
4A90
 
 | | Crystal structure of mouse SAP18 residues 1-143 | | Descriptor: | GLYCEROL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18 | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4NLJ
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P1) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
2X0G
 
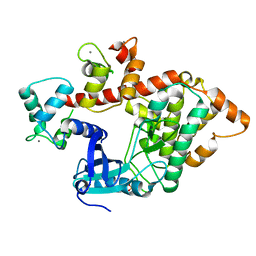 | | X-RAY STRUCTURE OF A DAP-KINASE CALMODULIN COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | Kuper, J, De Diego, I, Lehmann, F, Wilmanns, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of the Death-Associated Protein Kinase-Calcium/Calmodulin Regulator Complex.
Sci.Signal, 3, 2010
|
|
1Q6M
 
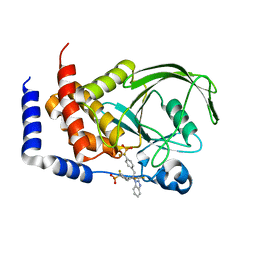 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 3 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, {[2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-(3,4-DIFLUOROPHENYL)PROPANE-1,3-DIYL]BIS[4,1-PHENYLENE(DIFLUOROMETHYLENE)]}BIS(PHOSPHONIC ACID) | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6P
 
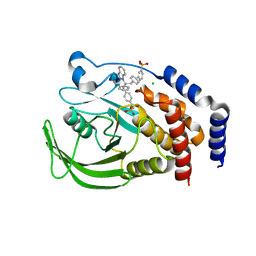 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 4'-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)-1,1'-BIPHENYL-3-YLPHOSPHONIC ACID, CHLORIDE ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6J
 
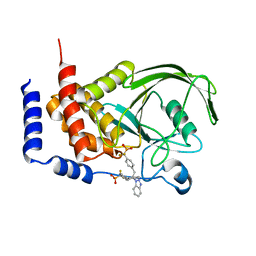 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 2 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, [4-(2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL](DIFLUORO)METHYLPHOSPHONIC ACID | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
4NLI
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P3121) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
1Q6T
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-[4-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-[(1S)-1-METHOXY-3-METHYLBUTYL]QUINOLIN-8-YLPHOSPHONIC ACID, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6S
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 9 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-((2R)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-METHYLQUINOLIN-8-YLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6N
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 4 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
9FPZ
 
 | | Human NatA-MAP2 80S ribosome complex | | Descriptor: | 28S rRNA (300-MER, fragments), 5.8S rRNA (58-MER), ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multi-protein assemblies orchestrate enzymatic processing of the nascent chain on the 80S ribosome
To Be Published
|
|
9ICD
 
 | | CATALYTIC MECHANISM OF NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE: IMPLICATIONS FROM THE STRUCTURES OF MAGNESIUM-ISOCITRATE AND NADP+ COMPLEXES | | Descriptor: | ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hurley, J.H, Dean, A.M, Koshland Jr, D.E, Stroud, R.M. | | Deposit date: | 1991-07-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes.
Biochemistry, 30, 1991
|
|
5AGH
 
 | | Crystal structure of the LeuRS editing domain of Candida albicans Mutant K510A | | Descriptor: | ACETATE ION, POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
1QZV
 
 | | Crystal structure of plant photosystem I | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Ben-Shem, A, Frolow, F, Nelson, N. | | Deposit date: | 2003-09-18 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.44 Å) | | Cite: | Crystal structure of plant photosystem I.
Nature, 426, 2003
|
|
9B8E
 
 | | Structure of S-nitrosylated Legionella pneumophila Ceg10. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Heisler, D.B, Alto, N.M. | | Deposit date: | 2024-03-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploiting bacterial effector proteins to uncover evolutionarily conserved antiviral host machinery.
Plos Pathog., 20, 2024
|
|
9XIM
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
6WGE
 
 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | Descriptor: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
9FQ0
 
 | | Human NatA-NAC-MAP1 80S ribosome complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Multi-protein assemblies orchestrate enzymatic processing of the nascent chain on the 80S ribosome
To Be Published
|
|
