19GS
 
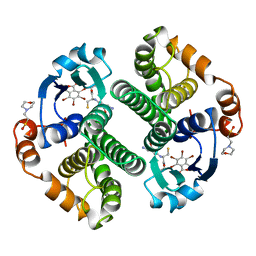 | | Glutathione s-transferase p1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-14 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1AH0
 
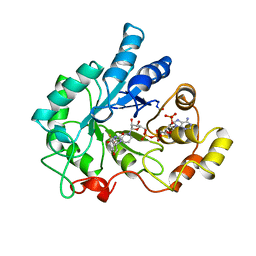 | | PIG ALDOSE REDUCTASE COMPLEXED WITH SORBINIL | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SORBINIL | | Authors: | Moras, D, Podjarny, A.D. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
1AH3
 
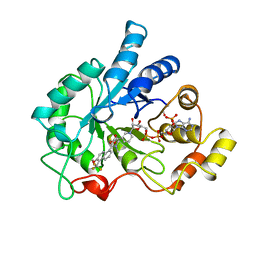 | | ALDOSE REDUCTASE COMPLEXED WITH TOLRESTAT INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Moras, D, Podjarny, A. | | Deposit date: | 1997-04-12 | | Release date: | 1998-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
5OY7
 
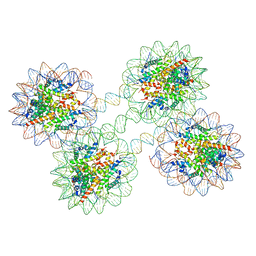 | | Structure of the 4_601_157 tetranucleosome (P1 form) | | Descriptor: | CHLORIDE ION, DNA (619-MER), Histone H2A, ... | | Authors: | Ekundayo, B, Richmond, T.J, Schalch, T. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.774 Å) | | Cite: | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
1D6W
 
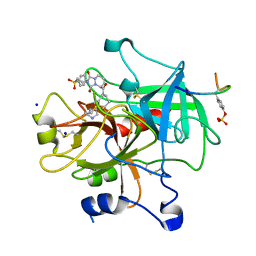 | | STRUCTURE OF THROMBIN COMPLEXED WITH SELECTIVE NON-ELECTROPHILIC INHIBITORS HAVING CYCLOHEXYL MOIETIES AT P1 | | Descriptor: | (5S)-N-[trans-4-(2-amino-1H-imidazol-5-yl)cyclohexyl]-1,3-dioxo-2-[2-(phenylsulfonyl)ethyl]-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazine-5-carboxamide, DECAPEPTIDE INHIBITOR, SODIUM ION, ... | | Authors: | Mochalkin, I, Tulinsky, A. | | Deposit date: | 1999-10-15 | | Release date: | 2000-10-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1L5W
 
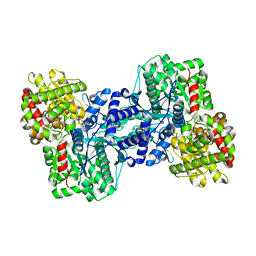 | | Crystal Structure of the Maltodextrin Phosphorylase Complexed with the Products of the Enzymatic Reaction between Glucose-1-phosphate and Maltotetraose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
2OI8
 
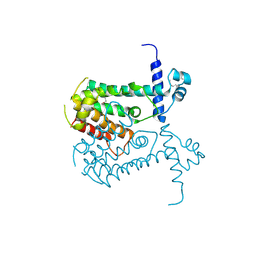 | | Crystal structure of putative regulatory protein SCO4313 | | Descriptor: | Putative regulatory protein SCO4313 | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tetR family protein SCO4313
To be Published
|
|
1D1W
 
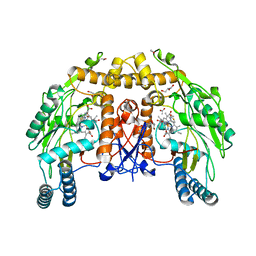 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 2-AMINOTHIAZOLINE (H4B BOUND) | | Descriptor: | 2-AMINOTHIAZOLINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-21 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
2BOF
 
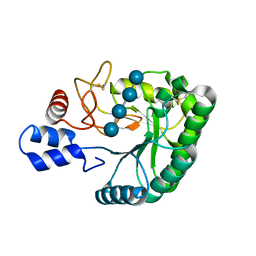 | | Catalytic domain of endo-1,4-glucanase Cel6A mutant Y73S from Thermobifida fusca in complex with cellotetrose | | Descriptor: | ENDOGLUCANASE E-2, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Larsson, A.M, Bergfors, T, Dultz, E, Irwin, D.C, Roos, A, Driguez, H, Wilson, D.B, Jones, T.A. | | Deposit date: | 2005-04-10 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Thermobifida Fusca Endoglucanase Cel6A in Complex with Substrate and Inhibitor: The Role of Tyrosine Y73 in Substrate Ring Distortion.
Biochemistry, 44, 2005
|
|
2BE3
 
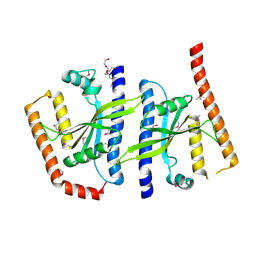 | |
2H26
 
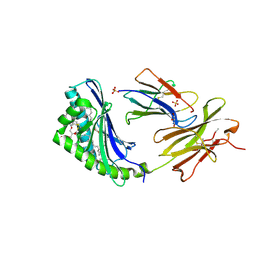 | | human CD1b in complex with endogenous phosphatidylcholine and spacer | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Vallina, A.T, Guillet, V, Mourey, L. | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b.
Embo J., 25, 2006
|
|
1QD1
 
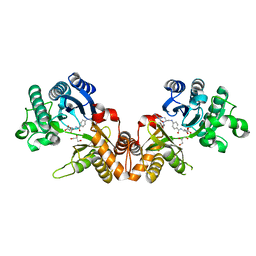 | | THE CRYSTAL STRUCTURE OF THE FORMIMINOTRANSFERASE DOMAIN OF FORMIMINOTRANSFERASE-CYCLODEAMINASE. | | Descriptor: | FORMIMINOTRANSFERASE-CYCLODEAMINASE, GLYCEROL, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid | | Authors: | Kohls, D, Sulea, T, Purisima, E, MacKenzie, R.E, Vrielink, A. | | Deposit date: | 1999-07-08 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the formiminotransferase domain of formiminotransferase-cyclodeaminase: implications for substrate channeling in a bifunctional enzyme.
Structure Fold.Des., 8, 2000
|
|
1BJ9
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, [7,12-DEACETYL-3,8,13,17-TETRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2-)-N21,N22,N23,N24]-IRON | | Authors: | Miller, M.A, Kraut, J. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
2OLB
 
 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
1NX6
 
 | |
1Q8A
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+:L-Hcy complex, Se-Met) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1DSA
 
 | | (+)-DUOCARMYCIN SA COVALENTLY LINKED TO DUPLEX DNA, NMR, 20 STRUCTURES | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Eis, P.S, Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA.
J.Mol.Biol., 272, 1997
|
|
1DM2
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR HYMENIALDISINE | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-AMINO-4-OXO-4H-PYRAZOL-3-YL)-2-BROMO-4,5,6,7-TETRAHYDRO-3AH-PYRROLO[2,3-C]AZEPIN-8-ONE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Thunnissen, A.M, Kim, S.-H. | | Deposit date: | 1999-12-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent.
Chem.Biol., 7, 2000
|
|
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1D9I
 
 | | STRUCTURE OF THROMBIN COMPLEXED WITH SELECTIVE NON-ELECTOPHILIC INHIBITORS HAVING CYCLOHEXYL MOIETIES AT P1 | | Descriptor: | (5S)-N-[(trans-4-aminocyclohexyl)methyl]-1,3-dioxo-2-[2-(phenylsulfonyl)ethyl]-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazine-5-carboxamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R, Tulinsky, A. | | Deposit date: | 1999-10-27 | | Release date: | 2000-10-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
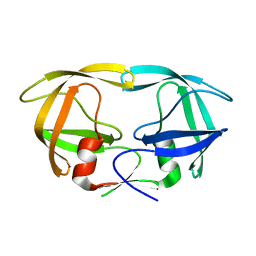
2HUB
 
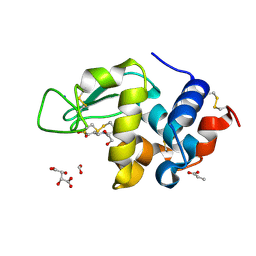 | | Structure of Hen Egg-White Lysozyme Determined from crystals grown in pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D(-)-TARTARIC ACID, ... | | Authors: | Frolow, F, Lagziel-Simis, S, Cohen-Hadar, N, Wine, Y, Freeman, A. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Monitoring influence of pH on the molecular and crystal structure of hen egg-white tetragonal lysozyme
To be Published
|
|
1LV1
 
 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1DSM
 
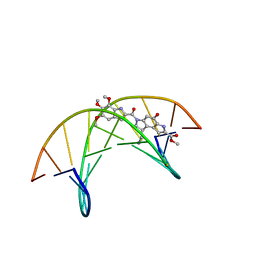 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
2A6U
 
 | |
