1T36
 
 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
1LXK
 
 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Tetrasaccharide Hyaluronan Substrate | | Descriptor: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | Deposit date: | 2002-06-05 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
1J71
 
 | |
7EHO
 
 | | Chitin oligosaccharide binding protein | | Descriptor: | Chitin oligosaccharide binding protein NagB2, TETRAETHYLENE GLYCOL | | Authors: | Itoh, T, Hibi, T, Kimoto, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of two solute-binding proteins for N,N' -diacetylchitobiose/ N,N',N'' -triacetylchitotoriose of the gram-positive bacterium, Paenibacillus sp. str. FPU-7.
J Struct Biol X, 5, 2021
|
|
7VPL
 
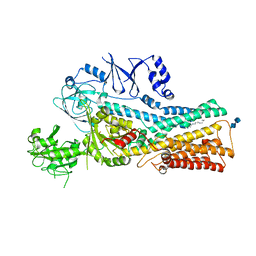 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
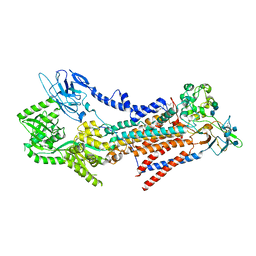
7VSH
 
 | |
7VPJ
 
 | | Cryo-EM structure of the human ATP13A2 (E1P-ADP state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
2LWZ
 
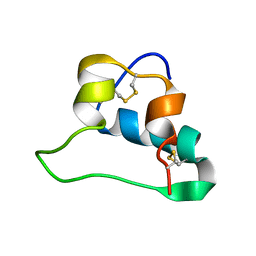 | | NMR Structures of Single-chain Insulin | | Descriptor: | SINGLE-CHAIN INSULIN | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2012-08-09 | | Release date: | 2013-08-28 | | Method: | SOLUTION NMR | | Cite: | Dynamic repair of an amyloidogenic protein: Insulin fibrillation is blocked by tethering a nascent alpha-helix
To be Published
|
|
5OT0
 
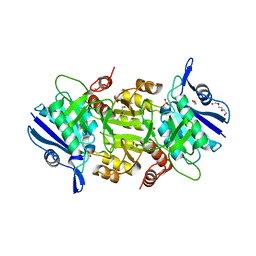 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7VGV
 
 | | Anion free form of light-driven chloride ion-pumping rhodopsin, NM-R3, structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, HEXADECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5OV3
 
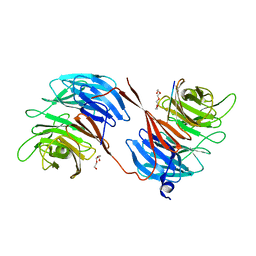 | | Structure of the RbBP5 beta-propeller domain | | Descriptor: | Retinoblastoma-binding protein 5, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Mittal, A, Zhang, Y, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2017-08-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the RbBP5 beta-propeller domain reveals a surface with potential nucleic acid binding sites.
Nucleic Acids Res., 46, 2018
|
|
7VW7
 
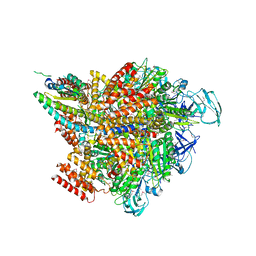 | | Crystal structure of the 2 ADP-AlF4-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Shekhar, M, Gupta, C, Singharoy, A, Murata, T. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.818 Å) | | Cite: | Revealing a Hidden Intermediate of Rotatory Catalysis with X-ray Crystallography and Molecular Simulations.
Acs Cent.Sci., 8, 2022
|
|
7V5S
 
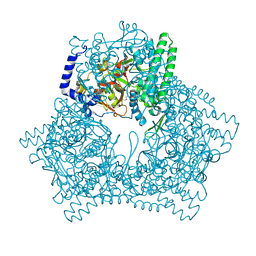 | | Crystal structure of human bleomycin hydrolase C73A mutant | | Descriptor: | Bleomycin hydrolase, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE4
 
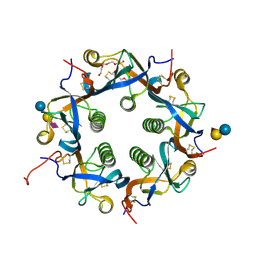 | | Crystal structure of Neu5Ac bound PltC | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE3
 
 | | Crystal structure of PltC | | Descriptor: | Subtilase cytotoxin subunit B-like protein, TETRAETHYLENE GLYCOL | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6A
 
 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6C
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6B
 
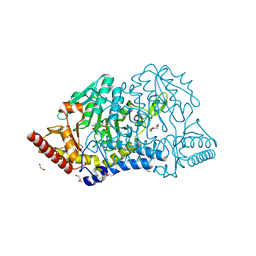 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7DU3
 
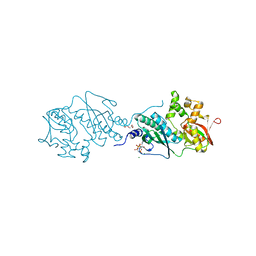 | | ThiL in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lin, J.Q, Lescar, J. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | ThiL in complex with AMP-PNP
To Be Published
|
|
7ESF
 
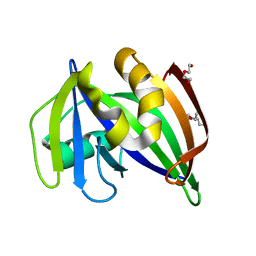 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7ET1
 
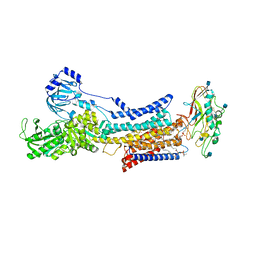 | |
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | Descriptor: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | Authors: | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
5QJ1
 
 | |
