7BSP
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in E1-AMPPCP state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BSV
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-occluded E2-AlF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, CDC50A, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | Authors: | Yan, X, Li, X, Song, G. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
7Y7U
 
 | |
7BSS
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in E1AlF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, CDC50A, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
6QZU
 
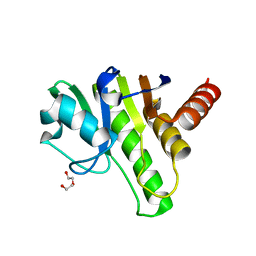 | | Getah virus macro domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0G
 
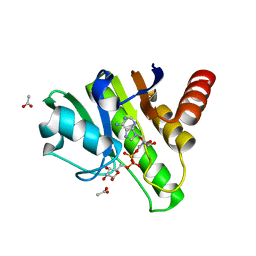 | | Getah virus macro domain in complex with ADPr, pose 2 | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural polyprotein | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0P
 
 | | Getah virus macro domain in complex with ADPr in double open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0F
 
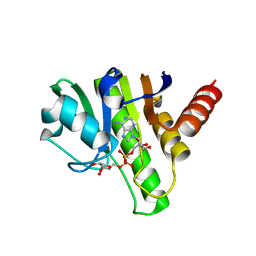 | | Getah virus macro domain in complex with ADPr, pose 1 | | Descriptor: | Non-structural polyprotein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0T
 
 | | Getah virus macro domain in complex with ADPr in open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0R
 
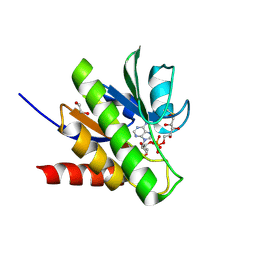 | | Getah virus macro domain in complex with ADPr covalently bond to Cys34 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural polyprotein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S})-2,3,4,5-tetrakis(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
4M0W
 
 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QID
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
5X7U
 
 | | Trehalose synthase from Thermobaculum terrenum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Trehalose synthase | | Authors: | Su, J, Wang, F. | | Deposit date: | 2017-02-27 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum.
J. Agric. Food Chem., 65, 2017
|
|
3ITG
 
 | |
3NVL
 
 | | Crystal Structure of Phosphoglycerate Mutase from Trypanosoma brucei | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, COBALT (II) ION, SULFATE ION | | Authors: | Mercaldi, G.F, Pereira, H.M, Cordeiro, A.T, Andricopulo, A.D, Thiemann, O.H. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphoglycerate mutase from Trypanosoma brucei.
Febs J., 279, 2012
|
|
3A4A
 
 | | Crystal structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase, alpha-D-glucopyranose | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
2IZA
 
 | | APOSTREPTAVIDIN PH 2.00 I4122 STRUCTURE | | Descriptor: | FORMIC ACID, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZJ
 
 | | STREPTAVIDIN-BIOTIN PH 3.50 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZI
 
 | | STREPTAVIDIN-BIOTIN PH 2.53 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
4LRU
 
 | | Crystal structure of glyoxalase III (Orf 19.251) from Candida albicans | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glyoxalase III (glutathione-independent) | | Authors: | Hasim, S, Hussin, N.A, Nickerson, K.W, Wilson, M.A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutathione-independent Glyoxalase of the DJ-1 Superfamily Plays an Important Role in Managing Metabolically Generated Methylglyoxal in Candida albicans.
J.Biol.Chem., 289, 2014
|
|
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
1XDB
 
 | | Crystal Structure of the Nitrogenase Fe protein Asp129Glu | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
3MJ5
 
 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | Descriptor: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | Authors: | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
