6KNZ
 
 | | Crystal structure of T2R-TTL-KXO1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[5-[4-(2-morpholin-4-ylethoxy)phenyl]pyridin-2-yl]-~{N}-(phenylmethyl)ethanamide, CALCIUM ION, ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.475 Å) | | Cite: | Reversible binding of the anticancer drug KXO1 (tirbanibulin) to the colchicine-binding site of beta-tubulin explains KXO1's low clinical toxicity.
J.Biol.Chem., 294, 2019
|
|
5SCU
 
 | |
6JT7
 
 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
6KAB
 
 | |
6KAY
 
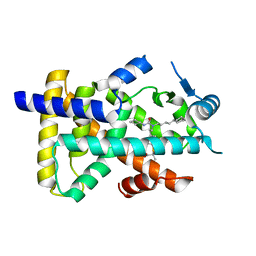 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5SCT
 
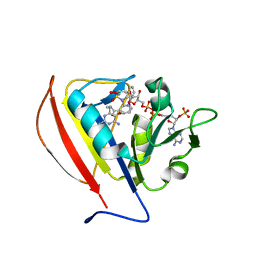 | |
6KB3
 
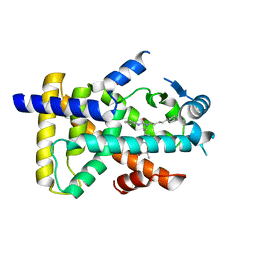 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5SD3
 
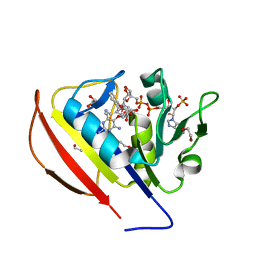 | |
6KBD
 
 | | fused To-MtbCsm1 with 2dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A),CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), MAGNESIUM ION | | Authors: | Li, T, Huo, Y, Jiang, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mycobacterium tuberculosis CRISPR/Cas system Csm1 holds clues to the evolutionary relationship between DNA polymerase and cyclase activity.
Int.J.Biol.Macromol., 170, 2020
|
|
5SCY
 
 | |
6JTO
 
 | | Crystal structure of HLA-C05 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational discovery of a cancer neoepitope harboring the KRAS G12D driver mutation.
Sci China Life Sci, 2021
|
|
6KCB
 
 | |
5SD0
 
 | |
6KCL
 
 | | Crystal structure of Plasmodium falciparum HPPK-DHPS A437G/K540E with pterin and p-hydroxybenzoate | | Descriptor: | 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
6JU4
 
 | | Aspergillus oryzae pro-tyrosinase F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUQ
 
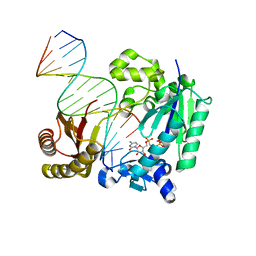 | | mutant PolIV-DNA incoming nucleotide complex 2 | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*CP*TP*GP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Johnson, M.K. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | A polar filter in DNA polymerases prevents ribonucleotide incorporation.
Nucleic Acids Res., 47, 2019
|
|
5SCS
 
 | |
6KD2
 
 | |
5SCZ
 
 | |
6KDH
 
 | | Antibody 64M-5 Fab including isoAsp in ligand-free form | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain) | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and biochemical basis of the formation of isoaspartate in the complementarity-determining region of antibody 64M-5 Fab.
Sci Rep, 9, 2019
|
|
6KDP
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, FORMIC ACID, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KE5
 
 | | Structure of CavAb in complex with Diltiazem and Amlodipine | | Descriptor: | CALCIUM ION, Ion transport protein, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Tang, L. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for Diltiazem Block of a Voltage-Gated Ca2+Channel.
Mol.Pharmacol., 96, 2019
|
|
6JV7
 
 | | Chimeric Rat C5a | | Descriptor: | Chimeric-rC5a, D-anaphylatoxin C5a, SULFATE ION | | Authors: | Zuo, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Chimeric protein probes for C5a receptors through fusion of anaphylatoxin C5a core region with a small-molecule antagonist
To Be Published
|
|
6JVH
 
 | | Crystal structure of human MTH1 in complex with compound MI0320 | | Descriptor: | 4-amino-6-fluoroquinoline-3-carbohydrazide, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
