9EO2
 
 | |
8ZUD
 
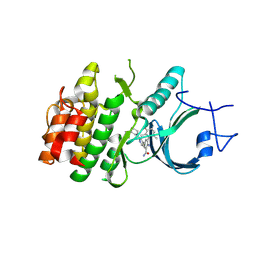 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8f | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-(2-morpholin-4-ylpyridin-4-yl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.50510085 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
9FR2
 
 | |
9FET
 
 | |
9C1B
 
 | | Crystal structure of GDP-bound human M-RAS protein in crystal form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9C1A
 
 | | Crystal structure of GDP-bound human M-RAS protein in crystal form I | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9BKF
 
 | |
9ENZ
 
 | |
9F4E
 
 | | UP1 in complex with Z1152242726 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-(6-methoxypyridin-3-yl)-N'-thiophen-2-ylurea | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-27 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4J
 
 | | UP1 in complex with Z416341642 | | Descriptor: | (2R)-N,2-dimethyl-N-(propan-2-yl)morpholine-4-sulfonamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4Q
 
 | | UP1 in complex with Z641239276 | | Descriptor: | 5-(2-phenylethylamino)pyrimidine-2,4-diol, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4N
 
 | | UP1 in complex with Z137811222 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4X
 
 | | UP1 in complex with Z1220452176 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F5C
 
 | | UP1 in complex with Z198195770 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-[3-(carbamoylamino)phenyl]acetamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F4V
 
 | | UP1 in complex with EN300-118084 | | Descriptor: | 3-(difluoromethyl)-1-methyl-1H-pyrazole-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F53
 
 | | UP1 in complex with Z237527902 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-(5-methyl-1H-pyrazol-3-yl)acetamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F52
 
 | | UP1 in complex with Z734147462 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, [4-(methylamino)oxan-4-yl]methanol | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9EO8
 
 | |
9J2N
 
 | | Cryo-EM structure of the human glucose transporter, GLUT7 in outward-facing open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 2, facilitated glucose transporter member 7 | | Authors: | Lee, S.S, Kim, S, Jin, M.S. | | Deposit date: | 2024-08-07 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human glucose transporter GLUT7.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
9J91
 
 | |
9RAT
 
 | |
9RSA
 
 | |
2BKT
 
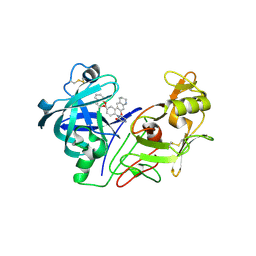 | | crystal structure of renin-pf00257567 complex | | Descriptor: | 1-{4-[3-(2-METHOXY-BENZYLOXY)-PROPOXY]-PHENYL}-6-(1,2,,3,4-TETRAHYDRO-QUINOLIN-7-YLOXYMETHYL)-PIPERAZIN-2-ONE, RENIN | | Authors: | Powell, N.A, Clay, E.H, Holsworth, D.D, Edmunds, J.J, Bryant, J.W, Ryan, J.M, Jalaie, M, Zhang, E. | | Deposit date: | 2005-02-18 | | Release date: | 2006-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Equipotent Activity in Both Enantiomers of a Series of Ketopiperazine-Based Renin Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7A49
 
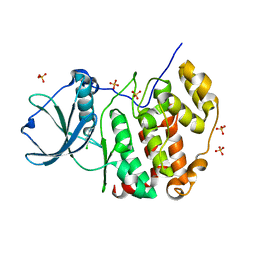 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6UVS
 
 | |
