6AMI
 
 | |
6UTR
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with copper | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
6M5M
 
 | | SPL-1 - GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetylglucosamine-specific lectin | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel carbohydrate-recognition mode of the invertebrate C-type lectin SPL-1 from Saxidomus purpuratus revealed by the GlcNAc-complex crystal in the presence of Ca2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5AX9
 
 | | Crystal structure of the kinase domain of human TRAF2 and NCK-interacting protein kinase in complex with compund 9 | | Descriptor: | 4-methoxy-3-[2-[(3-methoxy-4-morpholin-4-yl-phenyl)amino]pyridin-4-yl]benzenecarbonitrile, SULFATE ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
7TCS
 
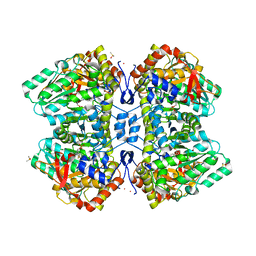 | | M379A mutant tyrosine phenol-lyase complexed with L-methionine | | Descriptor: | (4Z)-4-({[(1E)-1-carboxy-3-(methylsulfanyl)propylidene]azaniumyl}methylidene)-2-methyl-5-[(phosphonooxy)methyl]-1,4-dihydropyridin-3-olate, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | M379A Mutant Tyrosine Phenol-lyase from Citrobacter freundii Has Altered Conformational Dynamics.
Chembiochem, 23, 2022
|
|
5B0C
 
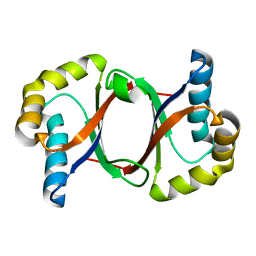 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
7T7N
 
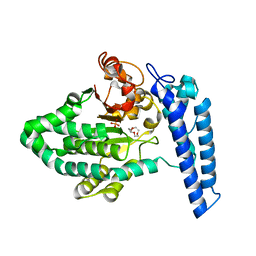 | | Structure of SPCC1393.13 protein from fission yeast | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Damage-control phosphatase SPCC1393.13, PHOSPHATE ION | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
5AP6
 
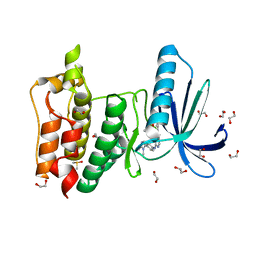 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.M, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
6OZ9
 
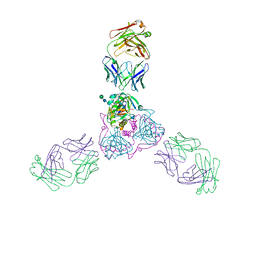 | | Ebola virus glycoprotein in complex with EBOV-520 Fab | | Descriptor: | EBOV-520 Fab heavy chain, EBOV-520 Fab light chain, Envelope glycoprotein, ... | | Authors: | Milligan, J.C, Altman, P.X, Hui, S, Hastie, K.M, Gilchuk, P, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.462 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6UO7
 
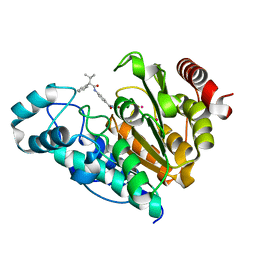 | |
8D0I
 
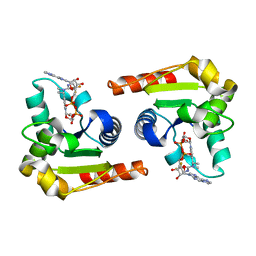 | | Human SARM1 bound to an NB-3 eADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
6UQ0
 
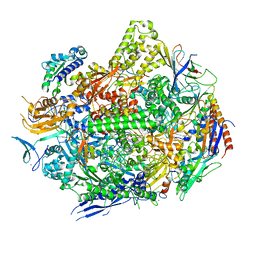 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 4 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
6UOE
 
 | |
5AQU
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1S,2R,3R,5R)-3-(HYDROXYMETHYL)-5-((5-METHOXYQUINAZOLIN-4-YL)AMINO)CYCLOPENTANE-1,2-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
6P2I
 
 | | Acyclic imino acid reductase (Bsp5) in complex with NADPH and D-Arg | | Descriptor: | 1,2-ETHANEDIOL, D-ARGININE, Glycerate dehydrogenase, ... | | Authors: | Guo, J, Higgins, M.A, Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An Asymmetric Reductase That Intercepts Acyclic Imino Acids Producedin Situby a Partner Oxidase.
J.Am.Chem.Soc., 141, 2019
|
|
7VA1
 
 | |
7TDL
 
 | | M379A mutant tyrosine phenol-lyase complexed with 3-bromo-DL-phenylalanine | | Descriptor: | (4Z)-4-({[(1E)-2-(3-bromophenyl)-1-carboxyethylidene]azaniumyl}methylidene)-2-methyl-5-[(phosphonooxy)methyl]-1,4-dihydropyridin-3-olate, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | M379A Mutant Tyrosine Phenol-lyase from Citrobacter freundii Has Altered Conformational Dynamics.
Chembiochem, 23, 2022
|
|
5AR0
 
 | | HSP72 with adenosine-derived inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(6-amino-8-((quinolin-7-ylmethyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cheeseman, M.D, Westwood, I.M, Barbeau, O, Rowlands, M.G, Jones, A.M, Jeganathan, F, Burke, R, Dobson, S.E, Workman, P, Collins, I, van Montfort, R.L.M, Jones, K. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of Hsp70.
J.Med.Chem., 59, 2016
|
|
7T7O
 
 | |
7T7K
 
 | | Structure of SPAC806.04c protein from fission yeast bound to Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Damage-control phosphatase SPAC806.04c, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
6AKQ
 
 | | The PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267 | | Descriptor: | PDZ tandem fragment of A. aeolicus site-2 protease homolog with the PA tag insertion | | Authors: | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
8D7F
 
 | | FlvF from Aspergillus flavus in complex with Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Terpene cyclase flvF | | Authors: | Tararina, M.A, Christianson, D.W. | | Deposit date: | 2022-06-07 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the Repurposed Fungal Terpene Cyclase FlvF Implicated in the C-N Bond-Forming Reaction of Flavunoidine Biosynthesis.
Biochemistry, 61, 2022
|
|
6UPD
 
 | | Structure of trehalose-6-phosphate phosphatase from Salmonella typhimurium in complex with trehalose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose-phosphate phosphatase, ... | | Authors: | Harvey, C.M, O'Toole, K.H, Allen, K.N. | | Deposit date: | 2019-10-17 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural Analysis of Binding Determinants ofSalmonella typhimuriumTrehalose-6-phosphate Phosphatase Using Ground-State Complexes.
Biochemistry, 59, 2020
|
|
6AL0
 
 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
