3HSP
 
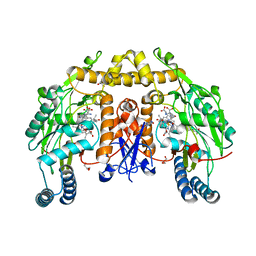 | | Ternary structure of neuronal nitric oxide synthase with NHA and NO bound(2) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Doukov, T, Li, H, Soltis, M, Poulos, T.L. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single crystal structural and absorption spectral characterizations of nitric oxide synthase complexed with N(omega)-hydroxy-L-arginine and diatomic ligands.
Biochemistry, 48, 2009
|
|
4D0T
 
 | | GalNAc-T2 crystal soaked with UDP-GalNAc, EA2 peptide and manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, MANGANESE (II) ION, ... | | Authors: | Lira-Navarrete, E, Iglesias-Fernandez, J, Zandberg, W.F, Companon, I, Kong, Y, Corzana, F, Pinto, B.M, Clausen, H, Peregrina, J.M, Vocadlo, D, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2014-04-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N-Acetylgalactosaminyltransferase 2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8EPC
 
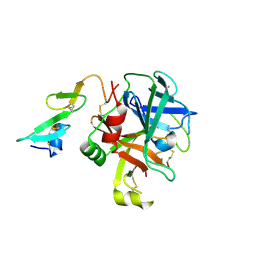 | |
3O1T
 
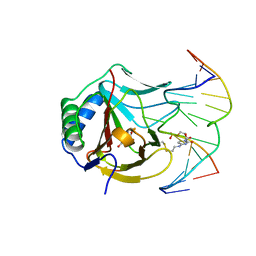 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*TP*AP*AP*(MDU)P*AP*CP*CP*GP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
8EPK
 
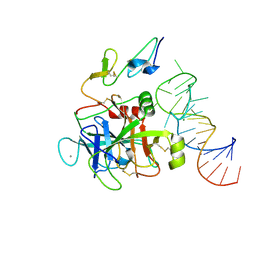 | |
3I2K
 
 | | Cocaine Esterase, wild type, bound to a DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
8EPH
 
 | | Crystal structure of human coagulation factor IXa (S195A), apo-form, DES-GLA | | Descriptor: | CALCIUM ION, Coagulation factor IXa heavy chain, Coagulation factor IXa light chain, ... | | Authors: | Kolyadko, V.N, Krishnaswamy, S. | | Deposit date: | 2022-10-05 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An RNA aptamer exploits exosite-dependent allostery to achieve specific inhibition of coagulation factor IXa.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6ME6
 
 | | XFEL crystal structure of human melatonin receptor MT2 in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
6SAM
 
 | | Structure of human butyrylcholinesterase in complex with 1-(2,3-dihydro-1H-inden2-yl)piperidin-3-yl N-phenyl carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Kosak, U, Strasek, N, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2019-07-17 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | N-alkylpiperidine carbamates as potential anti-Alzheimer's agents.
Eur.J.Med.Chem., 197, 2020
|
|
3SBR
 
 | | Pseudomonas stutzeri nitrous oxide reductase, P1 crystal form with substrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, DINUCLEAR COPPER ION, ... | | Authors: | Pomowski, A, Zumft, W.G, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | N2O binding at a [4Cu:2S] copper-sulphur cluster in nitrous oxide reductase.
Nature, 477, 2011
|
|
3I4D
 
 | | Photosynthetic reaction center from rhodobacter sphaeroides 2.4.1 | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Fujii, R, Adachi, S, Roszak, A.W, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W, Koshihara, S, Hashimoto, H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the carotenoid bound to the reaction centre from Rhodobacter sphaeroides 2.4.1 revealed by time-resolved X-ray crystallography
To be Published
|
|
5YFE
 
 | |
6MBC
 
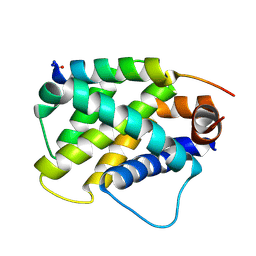 | | Human Bfl-1 in complex with the designed peptide dF4 | | Descriptor: | Bcl-2-related protein A1, dF4 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MG8
 
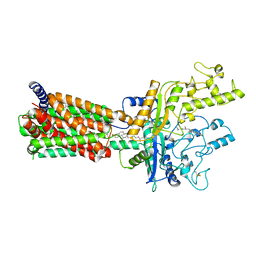 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
3HY8
 
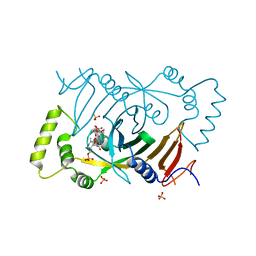 | | Crystal Structure of Human Pyridoxine 5'-Phosphate Oxidase R229W Mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Safo, M.K, Musayev, F.N, Di Salvo, M.L, Saavedra, M.K, Schirch, V. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of reduced pyridoxine 5'-phosphate oxidase catalytic activity in neonatal epileptic encephalopathy disorder
J.Biol.Chem., 284, 2009
|
|
6ME8
 
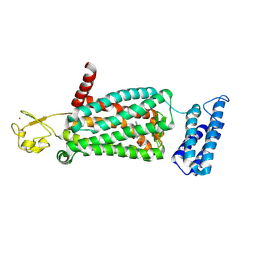 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
3VWI
 
 | | High resolution crystal structure of FraC in the monomeric form | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fragaceatoxin C, ... | | Authors: | Tanaka, K, Morante, K, Caaveiro, J.M.M, Gonzalez-Manas, J.M, Tsumoto, K. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
3NW2
 
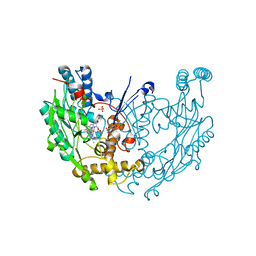 | | Novel nanomolar Imidazopyridines as selective Nitric Oxide Synthase (iNOS) inhibitors: SAR and structural insights | | Descriptor: | 2-[2-(4-methoxypyridin-2-yl)ethyl]-3H-imidazo[4,5-b]pyridine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Graedler, U, Fuchss, T, Ulrich, W.R, Boer, R, Strub, A, Hesslinger, C, Anezo, C, Diederichs, K, Zaliani, A. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel nanomolar imidazo[4,5-b]pyridines as selective nitric oxide synthase (iNOS) inhibitors: SAR and structural insights
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8F9Y
 
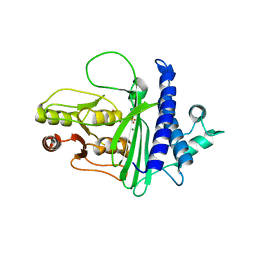 | | SAL1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SAL1 phosphatase | | Authors: | Frkic, R.L, Kaczmarski, J.A, Tan, L, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6MF8
 
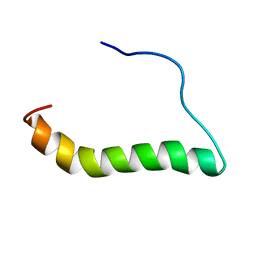 | | TCR alpha transmembrane domain | | Descriptor: | T-cell receptor alpha chain C region | | Authors: | Brazin, K.N, Reinherz, E.L. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The T Cell Antigen Receptor alpha Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations.
Immunity, 49, 2018
|
|
5YLV
 
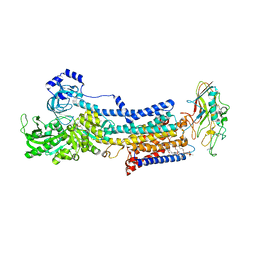 | | Crystal structure of the gastric proton pump complexed with SCH28080 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(2-methyl-8-phenylmethoxy-imidazo[1,2-a]pyridin-3-yl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79977775 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
3I0S
 
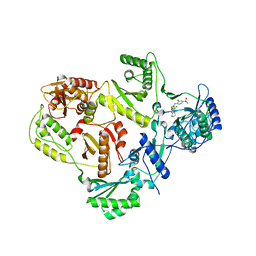 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 7 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6,8-dichloro-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6ME5
 
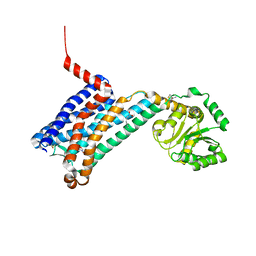 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6S8N
 
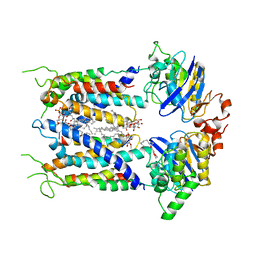 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
