2MC8
 
 | |
2MID
 
 | |
2MJ8
 
 | | Solution structure of CDYL2 chromodomain | | Descriptor: | Chromodomain Y-like protein 2 | | Authors: | Qin, S, Houliston, S, Arrowsmith, C.H, Edwards, A.M, Wu, H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-28 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2M9H
 
 | |
2MGY
 
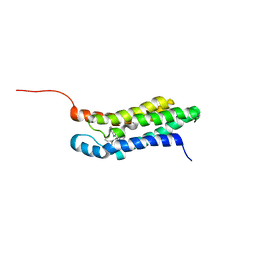 | | Solution structure of the mitochondrial translocator protein (TSPO) in complex with its high-affinity ligand PK11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the mitochondrial translocator protein in complex with a diagnostic ligand.
Science, 343, 2014
|
|
2ML1
 
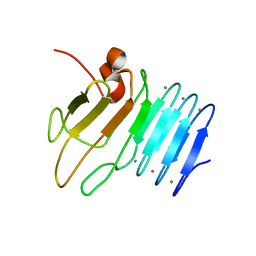 | |
2LXC
 
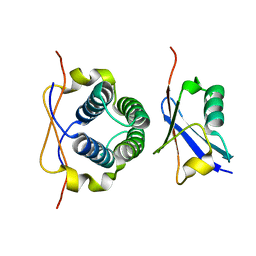 | |
2ME1
 
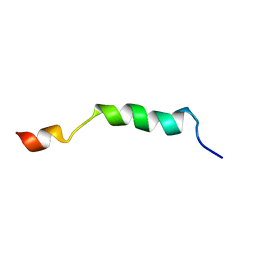 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2MGR
 
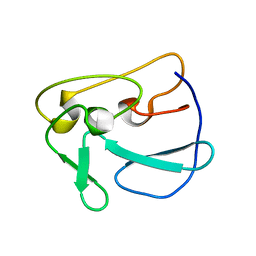 | | Structure of Plasmodium Yoelii Merozoite Surface Protein 1 - C-terminal Domain, E28K mutant | | Descriptor: | Merozoite surface protein 1 | | Authors: | Curd, R.D, Birdsall, B, Kadekoppala, M, Ogun, S, Kelly, G, Holder, A.A. | | Deposit date: | 2013-11-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of Plasmodium yoelii merozoite surface protein 119, antibody specificity and implications for malaria vaccine design
OPEN BIOLOGY, 4, 2014
|
|
2MKB
 
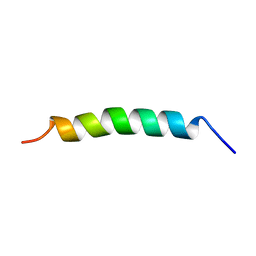 | |
2LYH
 
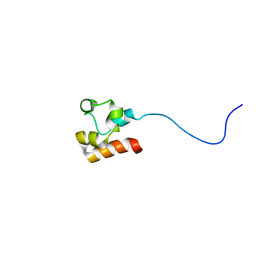 | | Structure of Faap24 residues 141-215 | | Descriptor: | Fanconi anemia-associated protein of 24 kDa | | Authors: | Wienk, H, Slootweg, J, Kaptein, R, Boelens, R, Folkers, G.E. | | Deposit date: | 2012-09-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Fanconi anemia associated protein FAAP24 uses two substrate specific binding surfaces for DNA recognition.
Nucleic Acids Res., 41, 2013
|
|
2MKL
 
 | |
2M3X
 
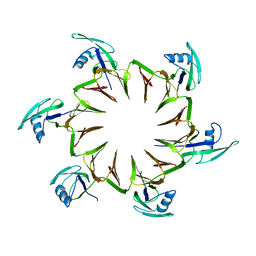 | |
2ML6
 
 | | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492 | | Descriptor: | Uncharacterized protein | | Authors: | Wallmann, A, Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-02-19 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492.
To be Published
|
|
2MMY
 
 | |
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2LX4
 
 | |
2LYB
 
 | |
2M9R
 
 | |
2MA2
 
 | | Solution structure of RasGRP2 EF hands bound to calcium | | Descriptor: | RAS guanyl-releasing protein 2 | | Authors: | Kuriyan, J, Iwig, J, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J, Wemmer, D, Roose, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
2M0M
 
 | | Structural Characterization of Minor Ampullate Spidroin Domains and their Distinct Roles in Fibroin Solubility and Fiber Formation | | Descriptor: | Minor ampullate fibroin 1 | | Authors: | Yang, D, Gao, Z, Lin, Z, Huang, W, Lai, C, Fan, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of minor ampullate spidroin domains and their distinct roles in fibroin solubility and fiber formation
Plos One, 8, 2013
|
|
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
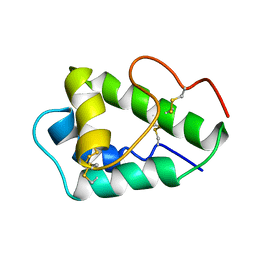
2MAL
 
 | |
2MAT
 
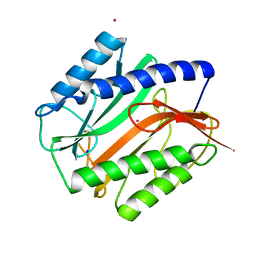 | | E.COLI METHIONINE AMINOPEPTIDASE AT 1.9 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2M6C
 
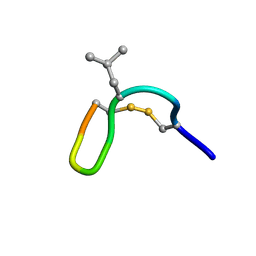 | | NMR solution structure of cis (minor) form of In936 in water | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-28 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
