3OE8
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
4A5Q
 
 | | Crystal structure of the chitinase Chi1 fitted into the 3D structure of the Yersinia entomophaga toxin complex | | Descriptor: | CHI1 | | Authors: | Busby, J.N, Landsberg, M.J, Simpson, R.M, Jones, S.A, Hankamer, B, Hurst, M.R.H, Lott, J.S. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-16 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structural Analysis of Chi1 Chitinase from Yen-Tc: The Multisubunit Insecticidal Abc Toxin Complex of Yersinia Entomophaga.
J.Mol.Biol., 415, 2012
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4AKH
 
 | | Dynein Motor Domain - AMPPNP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME, DYNEIN HEAVY CHAIN CYTOPLASMIC, ... | | Authors: | Schmidt, H, Gleave, E.S, Carter, A.P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Insights Into Dynein Motor Domain Function from a 3.3 Angstrom Crystal Structure
Nat.Struct.Mol.Biol., 19, 2012
|
|
3OJX
 
 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
4AQ9
 
 | |
4A2Y
 
 | | STRUCTURE OF THE HUMAN EOSINOPHIL CATIONIC PROTEIN IN COMPLEX WITH CITRATE ANIONS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Boix, E, Pulido, D, Moussaoui, M, Nogues, V, Russi, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Sulfate-Binding Site Structure of the Human Eosinophil Cationic Protein as Revealed by a New Crystal Form.
J.Struct.Biol., 179, 2012
|
|
3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4A2N
 
 | | Crystal Structure of Ma-ICMT | | Descriptor: | CARDIOLIPIN, ISOPRENYLCYSTEINE CARBOXYL METHYLTRANSFERASE, PALMITIC ACID, ... | | Authors: | Yang, J, Kulkarni, K, Manolaridis, I, Zhang, Z, Dodd, R.B, Mas-Droux, C, Barford, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Isoprenylcysteine Carboxyl Methylation from the Crystal Structure of the Integral Membrane Methyltransferase Icmt.
Mol.Cell, 44, 2011
|
|
4A6E
 
 | | Crystal structure of human N-acetylserotonin methyltransferase (ASMT) in complex with SAM and N-acetylserotonin | | Descriptor: | GLYCEROL, HYDROXYINDOLE O-METHYLTRANSFERASE, N-ACETYL SEROTONIN, ... | | Authors: | Legrand, P, Haouz, A, Shepard, W. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Mapping of Human Asmt, the Last Enzyme of the Melatonin Synthesis Pathway.
J.Pineal Res., 54, 2013
|
|
4A01
 
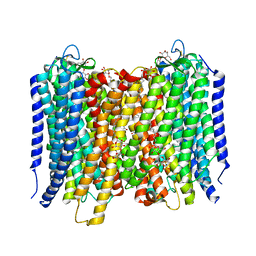 | | Crystal Structure of the H-Translocating Pyrophosphatase | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, ... | | Authors: | Lin, S.-M, Tsai, J.-Y, Hsiao, C.-D, Chiu, C.-L, Pan, R.-L, Sun, Y.-J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-28 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a Membrane Embedded H1-Translocating Pyrophosphatase
Nature, 484, 2012
|
|
3OMI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OJB
 
 | |
4A7Z
 
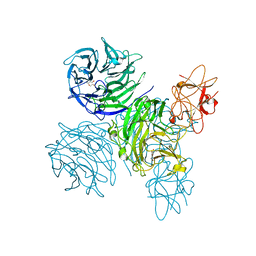 | | Complex of bifunctional aldos-2-ulose dehydratase with the reaction intermediate ascopyrone M | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, Ascopyrone M, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
3OJ6
 
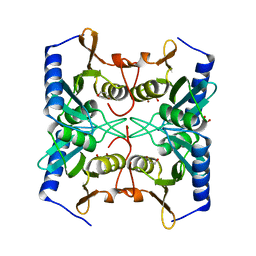 | |
3T0B
 
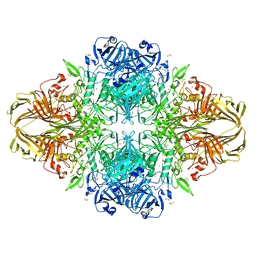 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
3SZG
 
 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
4A7Y
 
 | | Active site metal depleted aldos-2-ulose dehydratase | | Descriptor: | 1,5-anhydro-D-fructose, ALDOS-2-ULOSE DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4AE1
 
 | |
3T2T
 
 | |
3T1L
 
 | |
3T4U
 
 | |
3SRF
 
 | | Human M1 pyruvate kinase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
3SRH
 
 | | Human M2 pyruvate kinase | | Descriptor: | PHOSPHATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
