9DDE
 
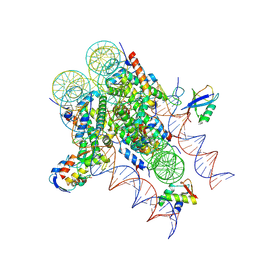 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
9DG3
 
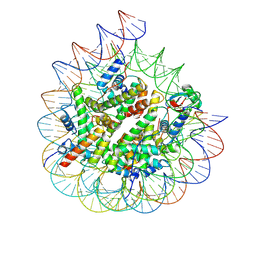 | | ncPRC1RYBP Delta-linker mutant bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-09-01 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
9DBY
 
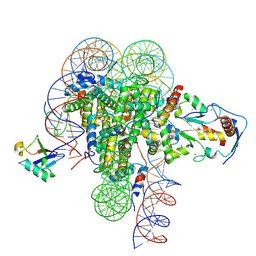 | | ncPRC1RYBP bound to singly modified H2AK119Ub nucleosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H2A type 1, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-24 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
7JOA
 
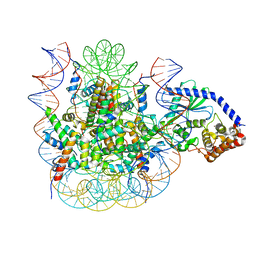 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
6Y5D
 
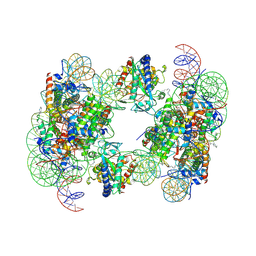 | | Structure of human cGAS (K394E) bound to the nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-A, ... | | Authors: | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
7JO9
 
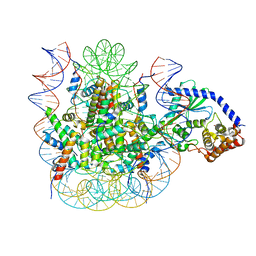 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
9B2T
 
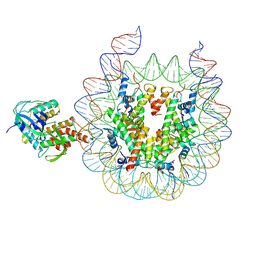 | | Haspin bound to nucleosome in position 2 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C. | | Deposit date: | 2024-03-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Haspin binds to nucleosomes using a unique DNA binding mechanism
To be published
|
|
9B2S
 
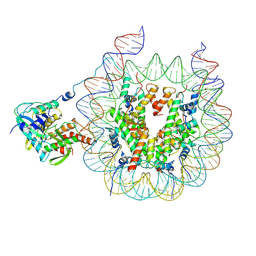 | | Haspin bound to nucleosome in position 1 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C. | | Deposit date: | 2024-03-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Haspin binds to nucleosomes using a unique DNA binding mechanism
To be published
|
|
6NE3
 
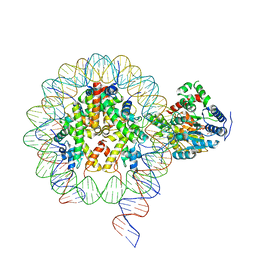 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | Authors: | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | Deposit date: | 2018-12-16 | | Release date: | 2019-07-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
9E1P
 
 | | Snf2h bound nucleosome complex - ClassB2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (152-MER), Histone H2A type 1, ... | | Authors: | Malik, D, Deshmukh, A.A, Bilokapic, S, Halic, M. | | Deposit date: | 2024-10-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Snf2h bound nucleosome complex-ClassB2
To Be Published
|
|
9E1Q
 
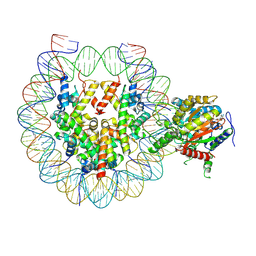 | | Snf2h bound nucleosome complex - ClassB3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (152-MER), Histone H2A type 1, ... | | Authors: | Malik, D, Deshmukh, A.A, Bilokapic, S, Halic, M. | | Deposit date: | 2024-10-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Snf2h bound nucleosome complex-ClassB3
To Be Published
|
|
9E1O
 
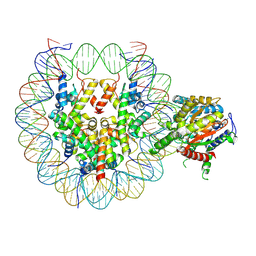 | | Snf2h bound nucleosome complex - ClassB1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (148-MER), DNA (150-MER), ... | | Authors: | Malik, D, Deshmukh, A.A, Bilokapic, S, Halic, M. | | Deposit date: | 2024-10-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snf2h bound nucleosome complex-ClassB1
To Be Published
|
|
9E1N
 
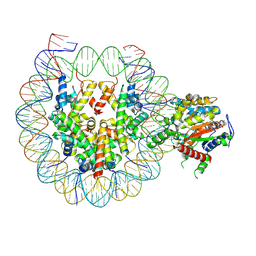 | | Snf2h bound nucleosome complex-ClassA3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (149-MER), DNA (151-MER), ... | | Authors: | Malik, D, Deshmukh, A.A, Bilokapic, S, Halic, M. | | Deposit date: | 2024-10-21 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Snf2h bound nucleosome complex-ClassA3
To Be Published
|
|
8PKJ
 
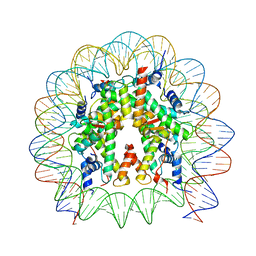 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8B0A
 
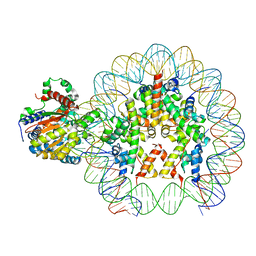 | | Cryo-EM structure of ALC1 bound to an asymmetric, site-specifically PARylated nucleosome | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1.
Nat Commun, 15, 2024
|
|
8G6G
 
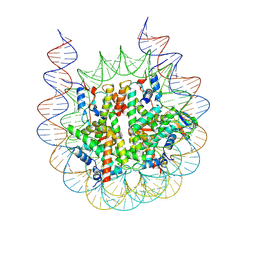 | | H2BK120ub+H3K79me2-modified nucleosome ubiquitin position 5 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
9GD0
 
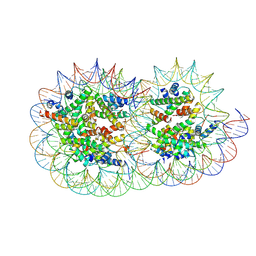 | | Structure of a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | DNA (250-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD2
 
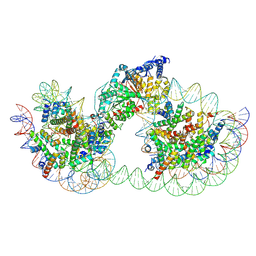 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
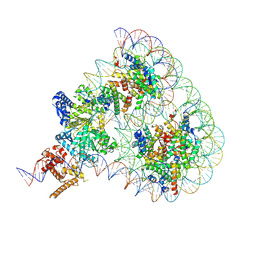 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD3
 
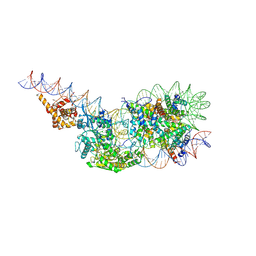 | | Structure of a mononucleosome bound by one copy of Chd1 with the DBD on the exit-side DNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
5KGF
 
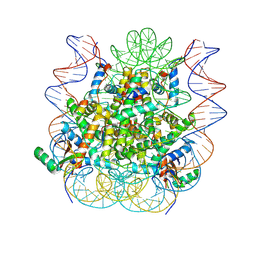 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
8CEO
 
 | |
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8BZ1
 
 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
