3EF0
 
 | |
3O32
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,5-dichlorobenzene-1,2-diol, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomytseva, M, Golovleva, L. | | Deposit date: | 2010-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3EMJ
 
 | |
3O5U
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
4CO2
 
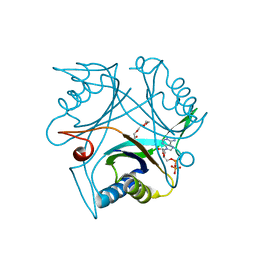 | |
2J78
 
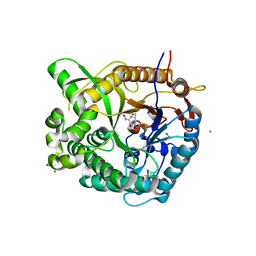 | | Beta-glucosidase from Thermotoga maritima in complex with gluco- hydroximolactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
4PA5
 
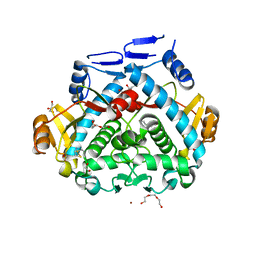 | | Tgl - a bacterial spore coat transglutaminase - cystamine complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, CITRATE ANION, ... | | Authors: | Brito, J.A, Placido, D, Fernandes, C.G, Lousa, D, Isidro, A, Soares, C.M, Pohl, J, Carrondo, M.A, Henriques, A.O, Archer, M. | | Deposit date: | 2014-04-07 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking.
Biochemistry, 54, 2015
|
|
4Q7Q
 
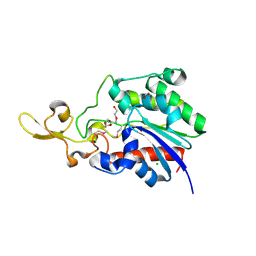 | | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Lipolytic protein G-D-S-L family, ... | | Authors: | Tan, K, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588
To be Published
|
|
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
3NUF
 
 | |
3FD3
 
 | | Structure of the C-terminal domains of a LysR family protein from Agrobacterium tumefaciens str. C58. | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, ... | | Authors: | Cuff, M.E, Xu, X, Zeng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-24 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the C-terminal domains of a LysR family protein from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
4D64
 
 | |
4HGA
 
 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GPQ
 
 | | Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Menin, ... | | Authors: | Shi, A, Murai, M.J, He, S, Lund, G.L, Hartley, T, Purohit, T, Reddy, G, Chruszcz, M, Grembecka, J, Cierpicki, T. | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia.
Blood, 120, 2012
|
|
4EE6
 
 | |
3RN1
 
 | |
4HOR
 
 | | Crystal Structure of Full-Length Human IFIT5 with 5`-triphosphate Oligocytidine | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 5, MAGNESIUM ION, RNA (5'-R(*(CTP)P*CP*CP*CP*C)-3') | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
4EC7
 
 | | Cobra NGF in complex with lipid | | Descriptor: | (2S)-1-hydroxy-3-(tetradecanoyloxy)propan-2-yl docosanoate, Venom nerve growth factor | | Authors: | Jiang, T, Wang, F, Tong, Q. | | Deposit date: | 2012-03-26 | | Release date: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into lipid-bound nerve growth factors
Faseb J., 26, 2012
|
|
3RMZ
 
 | |
4E8Y
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, D-beta-D-heptose 7-phosphate kinase, ... | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
3FES
 
 | | Crystal Structure of the ATP-dependent Clp Protease ClpC from Clostridium difficile | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp endopeptidase, MAGNESIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-01 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the ATP-dependent Clp Protease ClpC from Clostridium difficile
To be Published
|
|
4EE4
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with tetrasaccharide from Lacto-N-neohexose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
3FO3
 
 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium dithionite (sulfite complex) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2008-12-27 | | Release date: | 2009-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4HOU
 
 | | Crystal Structure of N-terminal Human IFIT1 | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1 | | Authors: | Abbas, Y.M, Pichlmair, A, Gorna, M.W, Superti-Furga, G, Nagar, B. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins.
Nature, 494, 2013
|
|
