1GLA
 
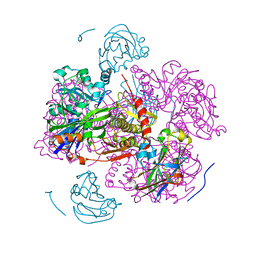 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, GLYCEROL KINASE | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
1EAA
 
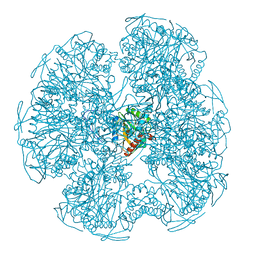 | |
5PAL
 
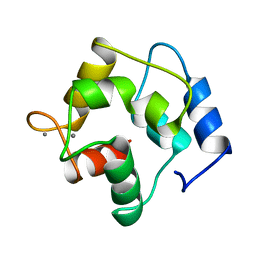 | | CRYSTAL STRUCTURE OF THE UNIQUE PARVALBUMIN COMPONENT FROM MUSCLE OF THE LEOPARD SHARK (TRIAKIS SEMIFASCIATA). THE FIRST X-RAY STUDY OF AN ALPHA-PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Roquet, F, Declercq, J.-P, Tinant, B, Rambaud, J, Parello, J. | | Deposit date: | 1991-09-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the unique parvalbumin component from muscle of the leopard shark (Triakis semifasciata). The first X-ray study of an alpha-parvalbumin.
J.Mol.Biol., 223, 1992
|
|
1AAN
 
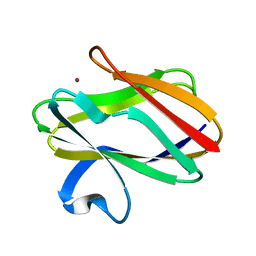 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
1EDE
 
 | |
3GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
1ALB
 
 | |
2SAS
 
 | |
1KDU
 
 | | SEQUENTIAL 1H NMR ASSIGNMENTS AND SECONDARY STRUCTURE OF THE KRINGLE DOMAIN FROM UROKINASE | | Descriptor: | PLASMINOGEN ACTIVATOR | | Authors: | Li, X, Bokman, A.M, Llinas, M, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kringle domain from urokinase-type plasminogen activator.
J.Mol.Biol., 235, 1994
|
|
1EAE
 
 | |
1L91
 
 | |
1L96
 
 | | STRUCTURE OF A HINGE-BENDING BACTERIOPHAGE T4 LYSOZYME MUTANT, ILE3-> PRO | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Dixon, M, Shewchuk, L, Matthews, B.W. | | Deposit date: | 1992-02-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile3-->Pro.
J.Mol.Biol., 227, 1992
|
|
1LFH
 
 | | MOLECULAR REPLACEMENT SOLUTION OF THE STRUCTURE OF APOLACTOFERRIN, A PROTEIN DISPLAYING LARGE-SCALE CONFORMATIONAL CHANGE | | Descriptor: | CHLORIDE ION, LACTOFERRIN | | Authors: | Anderson, B.F, Baker, E.N, Norris, G.E. | | Deposit date: | 1991-09-04 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular replacement solution of the structure of apolactoferrin, a protein displaying large-scale conformational change.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1C53
 
 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
1AFC
 
 | | STRUCTURAL STUDIES OF THE BINDING OF THE ANTI-ULCER DRUG SUCROSE OCTASULFATE TO ACIDIC FIBROBLAST GROWTH FACTOR | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Hsu, B.T, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the binding of the anti-ulcer drug sucrose octasulfate to acidic fibroblast growth factor.
Structure, 1, 1993
|
|
4FIS
 
 | | THE MOLECULAR STRUCTURE OF WILD-TYPE AND A MUTANT FIS PROTEIN: RELATIONSHIP BETWEEN MUTATIONAL CHANGES AND RECOMBINATIONAL ENHANCER FUNCTION OR DNA BINDING | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Yuan, H.S, Finkel, S.E, Feng, J.-A, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 1991-08-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1BTI
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | Authors: | Housset, D, Tao, F, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | Deposit date: | 1991-07-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
2TPR
 
 | |
1L87
 
 | |
1ADS
 
 | |
6CCP
 
 | |
1RDD
 
 | |
1AAL
 
 | |
1ABE
 
 | |
1NSC
 
 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
