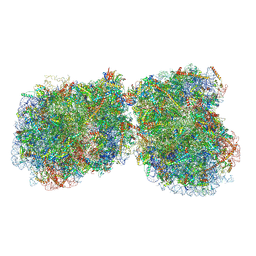
8Z9D
 
 | | cryo-EM structure of PSII-LHCII megacomplex from spinach | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J.Y, Liu, Z.F. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Architecture and functional regulation of a plant PSII-LHCII megacomplex.
Sci Adv, 10, 2024
|
|
8YK2
 
 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-700 | | Descriptor: | Alpha-galactosidase, GLYCEROL, SODIUM ION, ... | | Authors: | Kashima, T, Akama, M, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen.
J Appl Glycosci (1999), 71, 2024
|
|
6Q98
 
 | |
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLD
 
 | |
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q9A
 
 | |
6Q97
 
 | |
6Q95
 
 | |
8YMJ
 
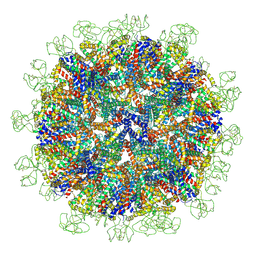 | | Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D2 symmetry | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
8ZM1
 
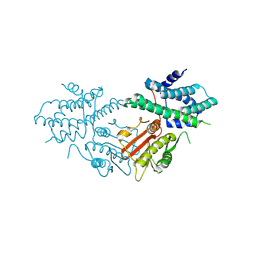 | | Structure of human pyruvate dehydrogenase kinase 2 complexed with compound 6 | | Descriptor: | (5~{R})-5-propan-2-ylindeno[1,2-b]pyridin-5-ol, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial | | Authors: | Akai, S, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-22 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and synthesis of novel fluorene derivatives as inhibitors of pyruvate dehydrogenase kinase.
Bioorg.Med.Chem.Lett., 109, 2024
|
|
8WQL
 
 | | In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | You, X, Zhang, X, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-10-11 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of in situ PBS-PSII supercomplex at 3.5 Angstroms resolution.
To Be Published
|
|
9B0Q
 
 | |
9B0S
 
 | |
3NSI
 
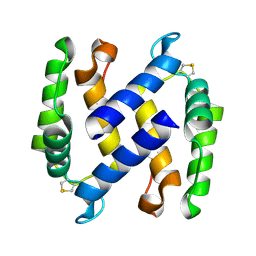 | |
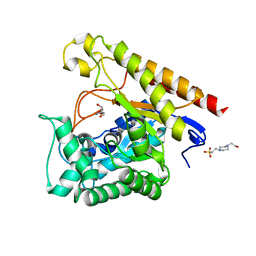
3NSK
 
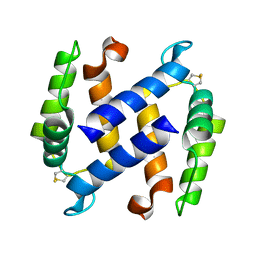 | |
4LYR
 
 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NV4
 
 | |
3NSL
 
 | |
4LYP
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NV2
 
 | |
3OKG
 
 | | Crystal structure of HsdS subunit from Thermoanaerobacter tengcongensis | | Descriptor: | Restriction endonuclease S subunits, SULFATE ION | | Authors: | Liang, D, Gao, P, Tang, Q, An, X, Yan, X. | | Deposit date: | 2010-08-24 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase
Plos One, 6, 2011
|
|
3OIT
 
 | | Crystal structure of curcuminoid synthase CUS from Oryza sativa | | Descriptor: | Os07g0271500 protein | | Authors: | Miyazono, K, Um, J, Imai, F.L, Katsuyama, Y, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of curcuminoid synthase CUS from Oryza sativa
Proteins, 79, 2011
|
|
